bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0310_orf1
Length=198
Score E
Sequences producing significant alignments: (Bits) Value
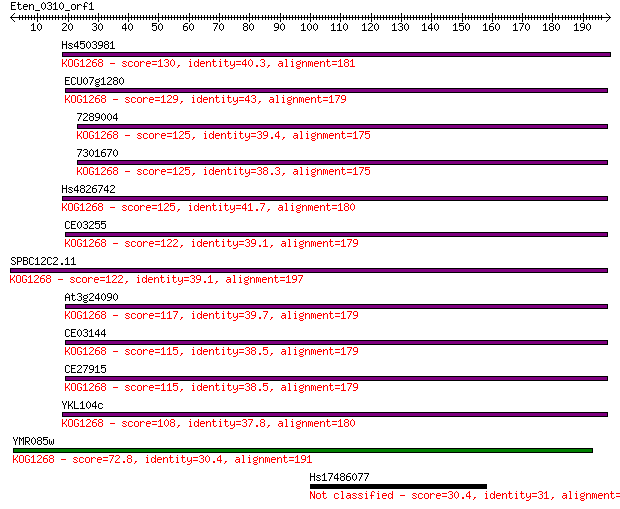
Hs4503981 130 2e-30
ECU07g1280 129 4e-30
7289004 125 6e-29
7301670 125 6e-29
Hs4826742 125 7e-29
CE03255 122 7e-28
SPBC12C2.11 122 7e-28
At3g24090 117 1e-26
CE03144 115 6e-26
CE27915 115 7e-26
YKL104c 108 1e-23
YMR085w 72.8 4e-13
Hs17486077 30.4 2.1
> Hs4503981
Length=681
Score = 130 bits (326), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 73/183 (39%), Positives = 108/183 (59%), Gaps = 4/183 (2%)
Query 18 DRCSALLDALHRLPVYAGMTLNAHGACKRIAEKLKDSQTLFVLGKGFGYPVALEGALKIK 77
+R ++ L RLP L+ +++A +L +++ ++G+G+ Y LEGALKIK
Sbjct 501 ERRKEIMLGLKRLPDLIKEVLSMDDEIQKLATELYHQKSVLIMGRGYHYATCLEGALKIK 560
Query 78 EISYIHAEGFAGGALKHGPFALIDDKEQTPVIMIVLGDQHAALMLNAAQQVKARGAY--L 135
EI+Y+H+EG G LKHGP AL+D + PVIMI++ D A NA QQV AR +
Sbjct 561 EITYMHSEGILAGELKHGPLALVD--KLMPVIMIIMRDHTYAKCQNALQQVVARQGRPVV 618
Query 136 ICLTDFPELVQDVADDYIVIPSNGPLTALLAAVPLQLLAYELSIARGINPDKPRGLAKTV 195
IC + E +++ V S L +L+ +PLQLLA+ L++ RG + D PR LAK+V
Sbjct 619 ICDKEDTETIKNTKRTIKVPHSVDCLQGILSVIPLQLLAFHLAVLRGYDVDFPRNLAKSV 678
Query 196 TVS 198
TV
Sbjct 679 TVE 681
> ECU07g1280
Length=699
Score = 129 bits (324), Expect = 4e-30, Method: Compositional matrix adjust.
Identities = 77/184 (41%), Positives = 107/184 (58%), Gaps = 10/184 (5%)
Query 19 RCSALLDALHRLPVYAGMTLNAHGACKRIAEK-LKDSQTLFVLGKGFGYPVALEGALKIK 77
R +++ L + L + K +A +KD +L ++G+G+ YP +EGALKIK
Sbjct 520 RRREIMEGLKNISSQINRVLELSTSVKSLANGPMKDDASLLLIGRGYQYPTCMEGALKIK 579
Query 78 EISYIHAEGFAGGALKHGPFALIDDKEQTPVIMIVLGDQHAALMLNAAQQVKARGAY--L 135
EI+YIHAEG A G LKHGP AL+DDK + +I I D NA +Q+ ARG +
Sbjct 580 EITYIHAEGLAAGELKHGPIALVDDKLR--IIFIATKDLLYDKTRNAMEQIFARGGRPIV 637
Query 136 ICLTDFPELVQDVAD-DYIVIPSN-GPLTALLAAVPLQLLAYELSIARGINPDKPRGLAK 193
IC D + D A+ D V+P L +L +PLQLL+Y L++A+G N D PR LAK
Sbjct 638 ICTED---ISGDYAEYDTFVVPKTVDCLQGILTVIPLQLLSYHLAVAKGYNADFPRNLAK 694
Query 194 TVTV 197
+VTV
Sbjct 695 SVTV 698
> 7289004
Length=694
Score = 125 bits (314), Expect = 6e-29, Method: Compositional matrix adjust.
Identities = 69/177 (38%), Positives = 104/177 (58%), Gaps = 4/177 (2%)
Query 23 LLDALHRLPVYAGMTLNAHGACKRIAEKLKDSQTLFVLGKGFGYPVALEGALKIKEISYI 82
+L AL +L L K +A+ L ++L ++G+G+ + LEGALK+KE++Y+
Sbjct 519 ILQALSKLADQIRDVLQLDSKVKELAKDLYQHKSLLIMGRGYNFATCLEGALKVKELTYM 578
Query 83 HAEGFAGGALKHGPFALIDDKEQTPVIMIVLGDQHAALMLNAAQQVKAR-GAYLICLTDF 141
H+EG G LKHGP AL+DD PV+MIVL D +NA QQV +R G +I +
Sbjct 579 HSEGIMAGELKHGPLALVDD--SMPVLMIVLRDPVYVKCMNALQQVTSRKGCPIIICEEG 636
Query 142 PELVQDVADDYIVIPSN-GPLTALLAAVPLQLLAYELSIARGINPDKPRGLAKTVTV 197
E + + ++ IP L +L +P+QLL+Y +++ RG + D PR LAK+VTV
Sbjct 637 DEETKAFSSRHLEIPRTVDCLQGILTVIPMQLLSYHIAVLRGCDVDCPRNLAKSVTV 693
> 7301670
Length=683
Score = 125 bits (313), Expect = 6e-29, Method: Compositional matrix adjust.
Identities = 67/177 (37%), Positives = 106/177 (59%), Gaps = 4/177 (2%)
Query 23 LLDALHRLPVYAGMTLNAHGACKRIAEKLKDSQTLFVLGKGFGYPVALEGALKIKEISYI 82
++D L +L + L + +++A++L + ++L ++G+GF + LEGALK+KE++Y+
Sbjct 508 IIDGLSQLDEHIRTVLKLNSQVQQLAKELYEHKSLLIMGRGFNFATCLEGALKVKELTYM 567
Query 83 HAEGFAGGALKHGPFALIDDKEQTPVIMIVLGDQHAALMLNAAQQVKARGAY--LICLTD 140
H+EG G LKHGP AL+D ++ PV+MIVL D +NA QQV +R LIC
Sbjct 568 HSEGILAGELKHGPLALVD--KEMPVLMIVLRDPVYTKCMNALQQVTSRKGRPILICEEG 625
Query 141 FPELVQDVADDYIVIPSNGPLTALLAAVPLQLLAYELSIARGINPDKPRGLAKTVTV 197
E + + + L +L +PLQLL+Y +++ RG + D PR LAK+VTV
Sbjct 626 DNETMSFSTRSLQIPRTVDCLQGVLTVIPLQLLSYHIAVLRGCDVDCPRNLAKSVTV 682
> Hs4826742
Length=682
Score = 125 bits (313), Expect = 7e-29, Method: Compositional matrix adjust.
Identities = 75/183 (40%), Positives = 105/183 (57%), Gaps = 6/183 (3%)
Query 18 DRCSALLDALHRLPVYAGMTLNAHGACKRIAEKLKDSQTLFVLGKGFGYPVALEGALKIK 77
+R ++ L LP L+ +A +L ++L V+G+G+ Y LEGALKIK
Sbjct 502 NRRQEIIRGLRSLPELIKEVLSLEEKIHDLALELYTQRSLLVMGRGYNYATCLEGALKIK 561
Query 78 EISYIHAEGFAGGALKHGPFALIDDKEQTPVIMIVLGDQHAALMLNAAQQVKARGAY--L 135
EI+Y+H+EG G LKHGP ALID +Q PVIM+++ D A NA QQV AR +
Sbjct 562 EITYMHSEGILAGELKHGPLALID--KQMPVIMVIMKDPCFAKCQNALQQVTARQGRPII 619
Query 136 ICLTDFPELVQDVADDYIVIPSN-GPLTALLAAVPLQLLAYELSIARGINPDKPRGLAKT 194
+C D E A I +P L +L+ +PLQLL++ L++ RG + D PR LAK+
Sbjct 620 LCSKDDTE-SSKFAYKTIELPHTVDCLQGILSVIPLQLLSFHLAVLRGYDVDFPRNLAKS 678
Query 195 VTV 197
VTV
Sbjct 679 VTV 681
> CE03255
Length=710
Score = 122 bits (305), Expect = 7e-28, Method: Compositional matrix adjust.
Identities = 70/182 (38%), Positives = 103/182 (56%), Gaps = 5/182 (2%)
Query 19 RCSALLDALHRLPVYAGMTLNAHGACKRIAEKLKDSQTLFVLGKGFGYPVALEGALKIKE 78
R + ++DAL+ LP+ L+ +IAE++ ++L ++G+G + LEGALKIKE
Sbjct 530 RRAEIIDALNNLPILIRDVLDLDDQVLKIAEQIYKDKSLLIMGRGLNFATCLEGALKIKE 589
Query 79 ISYIHAEGFAGGALKHGPFALIDDKEQTPVIMIVLGDQHAALMLNAAQQVKARGAYLICL 138
+SY+H EG G LKHGP A++D E + M+V D LNA QQV AR I +
Sbjct 590 LSYMHCEGIMSGELKHGPLAMVD--EFLSICMVVCNDHVYKKSLNALQQVVARKGAPIII 647
Query 139 TDFPELVQDVA--DDYIVIPSN-GPLTALLAAVPLQLLAYELSIARGINPDKPRGLAKTV 195
D D+A + +P + +L +PLQLL+Y ++ G N D+PR LAK+V
Sbjct 648 ADSSVPESDLAGMKHVLRVPRTVDCVQNILTVIPLQLLSYHIAELNGANVDRPRNLAKSV 707
Query 196 TV 197
TV
Sbjct 708 TV 709
> SPBC12C2.11
Length=696
Score = 122 bits (305), Expect = 7e-28, Method: Compositional matrix adjust.
Identities = 77/199 (38%), Positives = 114/199 (57%), Gaps = 5/199 (2%)
Query 1 ALVASWFWQNLRSEEYPDRCSALLDALHRLPVYAGMTLNAHGACKRIA-EKLKDSQTLFV 59
ALV + + S +R + ++D L + TL+ + A K+ A E+L + + +
Sbjct 500 ALVLMALYLSRDSVSRLERRNEIIDGLAEIGEKVQETLHLNAAIKQTAIEQLINKDKMLI 559
Query 60 LGKGFGYPVALEGALKIKEISYIHAEGFAGGALKHGPFALIDDKEQTPVIMIVLGDQHAA 119
+G+G+ Y ALEGALK+KEISY HAEG G LKHG AL+D+ P++M++ D +
Sbjct 560 IGRGYHYATALEGALKVKEISYTHAEGVLAGELKHGVLALVDN--DMPIVMLLPDDYNFP 617
Query 120 LMLNAAQQVKARGAYLICLTDFPELVQDVADDYIVIPSN-GPLTALLAAVPLQLLAYELS 178
NA +QV+ARG I +TD +L I +P L +L +P QLL+Y L+
Sbjct 618 KAWNAFEQVRARGGKPIIITD-KKLDNLEGFTIIKVPKTVDCLQGILNVIPFQLLSYWLA 676
Query 179 IARGINPDKPRGLAKTVTV 197
+ RG N D+PR LAK+VTV
Sbjct 677 VKRGHNVDQPRNLAKSVTV 695
> At3g24090
Length=691
Score = 117 bits (293), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 71/183 (38%), Positives = 100/183 (54%), Gaps = 6/183 (3%)
Query 19 RCSALLDALHRLPVYAGMTLNAHGACKRIAEKLKDSQTLFVLGKGFGYPVALEGALKIKE 78
R A++D L LP L K +A+ L D Q+L V G+G+ Y ALEGALK+KE
Sbjct 510 RREAIIDGLLDLPYKVKEVLKLDDEMKDLAQLLIDEQSLLVFGRGYNYATALEGALKVKE 569
Query 79 ISYIHAEGFAGGALKHGPFALIDDKEQTPVIMIVLGDQHAALMLNAAQQVKARGAYLICL 138
++ +H+EG G +KHGP AL+D E P+ +I D + + QQ+ AR LI +
Sbjct 570 VALMHSEGILAGEMKHGPLALVD--ENLPIAVIATRDACFSKQQSVIQQLHARKGRLIVM 627
Query 139 T---DFPELVQDVADDYIVIPS-NGPLTALLAAVPLQLLAYELSIARGINPDKPRGLAKT 194
D + + I +P L ++ VPLQLLAY L++ RG N D+PR LAK+
Sbjct 628 CSKGDAASVSSSGSCRAIEVPQVEDCLQPVINIVPLQLLAYHLTVLRGHNVDQPRNLAKS 687
Query 195 VTV 197
VT
Sbjct 688 VTT 690
> CE03144
Length=725
Score = 115 bits (288), Expect = 6e-26, Method: Compositional matrix adjust.
Identities = 69/182 (37%), Positives = 100/182 (54%), Gaps = 5/182 (2%)
Query 19 RCSALLDALHRLPVYAGMTLNAHGACKRIAEKLKDSQTLFVLGKGFGYPVALEGALKIKE 78
R ++DAL+ LP L IA+++ ++L ++G+G + LEGALKIKE
Sbjct 545 RREEIIDALNDLPELIREVLQLDEKVLDIAKQIYKEKSLLIMGRGLNFATCLEGALKIKE 604
Query 79 ISYIHAEGFAGGALKHGPFALIDDKEQTPVIMIVLGDQHAALMLNAAQQVKARGAYLICL 138
+SY+H EG G LKHGP A++D E + M+V D+ LNA QQV AR I +
Sbjct 605 LSYMHCEGIMSGELKHGPLAMVD--EFLSICMVVCNDRVYKKSLNALQQVVARKGAPIII 662
Query 139 TDFPELVQDVA--DDYIVIPSN-GPLTALLAAVPLQLLAYELSIARGINPDKPRGLAKTV 195
D D+A + +P + +L +PLQLL+Y ++ G N D+PR LAK+V
Sbjct 663 ADCTVPEGDLAGMKHILRVPKTVDCVQNILTVIPLQLLSYHIAELNGANVDRPRNLAKSV 722
Query 196 TV 197
TV
Sbjct 723 TV 724
> CE27915
Length=712
Score = 115 bits (287), Expect = 7e-26, Method: Compositional matrix adjust.
Identities = 69/182 (37%), Positives = 100/182 (54%), Gaps = 5/182 (2%)
Query 19 RCSALLDALHRLPVYAGMTLNAHGACKRIAEKLKDSQTLFVLGKGFGYPVALEGALKIKE 78
R ++DAL+ LP L IA+++ ++L ++G+G + LEGALKIKE
Sbjct 532 RREEIIDALNDLPELIREVLQLDEKVLDIAKQIYKEKSLLIMGRGLNFATCLEGALKIKE 591
Query 79 ISYIHAEGFAGGALKHGPFALIDDKEQTPVIMIVLGDQHAALMLNAAQQVKARGAYLICL 138
+SY+H EG G LKHGP A++D E + M+V D+ LNA QQV AR I +
Sbjct 592 LSYMHCEGIMSGELKHGPLAMVD--EFLSICMVVCNDRVYKKSLNALQQVVARKGAPIII 649
Query 139 TDFPELVQDVA--DDYIVIPSN-GPLTALLAAVPLQLLAYELSIARGINPDKPRGLAKTV 195
D D+A + +P + +L +PLQLL+Y ++ G N D+PR LAK+V
Sbjct 650 ADCTVPEGDLAGMKHILRVPKTVDCVQNILTVIPLQLLSYHIAELNGANVDRPRNLAKSV 709
Query 196 TV 197
TV
Sbjct 710 TV 711
> YKL104c
Length=717
Score = 108 bits (269), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 68/187 (36%), Positives = 106/187 (56%), Gaps = 9/187 (4%)
Query 18 DRCSALLDALHRLPVYAGMTLNAHGACKRI-AEKLKDSQTLFVLGKGFGYPVALEGALKI 76
DR ++ L +P L K++ A +LKD ++L +LG+G+ + ALEGALKI
Sbjct 532 DRRIEIIQGLKLIPGQIKQVLKLEPRIKKLCATELKDQKSLLLLGRGYQFAAALEGALKI 591
Query 77 KEISYIHAEGFAGGALKHGPFALIDDKEQTPVIMIVLGDQHAALMLNAAQQVKARGAYLI 136
KEISY+H+EG G LKHG AL+D E P+I D ++++ +QV AR + I
Sbjct 592 KEISYMHSEGVLAGELKHGVLALVD--ENLPIIAFGTRDSLFPKVVSSIEQVTARKGHPI 649
Query 137 CLTDFPELV-----QDVADDYIVIPSN-GPLTALLAAVPLQLLAYELSIARGINPDKPRG 190
+ + + V + + + +P L L+ +PLQL++Y L++ +GI+ D PR
Sbjct 650 IICNENDEVWAQKSKSIDLQTLEVPQTVDCLQGLINIIPLQLMSYWLAVNKGIDVDFPRN 709
Query 191 LAKTVTV 197
LAK+VTV
Sbjct 710 LAKSVTV 716
> YMR085w
Length=432
Score = 72.8 bits (177), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 58/198 (29%), Positives = 91/198 (45%), Gaps = 11/198 (5%)
Query 2 LVASWFWQNLRSEEYPDRCSALLDALHRLPVYAGMTLNAHGACKRIAEK-LKDSQTLFVL 60
++A W ++L S+ +R ++ AL +P L + +K LK T +L
Sbjct 233 MIALWMSEDLVSK--IERRKEIIQALTIVPSQIKEVLELEPLIIELCDKKLKQHDTFLLL 290
Query 61 GKGFGYPVALEGALKIKEISYIHAEGFAGGALKHGPFALIDDKEQTPVIMIVLGDQHAAL 120
G+G+ + ALEGA K+KEISY+H+E L H A+ D P+I D +
Sbjct 291 GRGYQFASALEGASKMKEISYVHSESILTNELGHRVLAVASD--NPPIIAFATKDAFSPK 348
Query 121 MLNAAQQVKAR--GAYLICLTDFPELVQDVADDYIV---IPSN-GPLTALLAAVPLQLLA 174
+ + Q+ R +IC QD +V +P L +L +PLQL++
Sbjct 349 IASCIDQIIERKGNPIIICNKGHKIWEQDKQKGNVVTLEVPQTVDCLQGILNVIPLQLIS 408
Query 175 YELSIARGINPDKPRGLA 192
Y L+I + I D PR A
Sbjct 409 YWLAIKKDIGVDLPRDSA 426
> Hs17486077
Length=399
Score = 30.4 bits (67), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 18/70 (25%), Positives = 35/70 (50%), Gaps = 12/70 (17%)
Query 100 IDDKEQTPVIMIVLGDQHAALMLNAAQQVKAR---------GAYLICLTDFPEL---VQD 147
+DDK +++ QH A LNA Q+++ + G +C+ + P + D
Sbjct 330 VDDKYDRMKTCLLMRQQHEAAALNAVQRMEWQLKVQELDPAGHKSLCVNEVPSFYVPMVD 389
Query 148 VADDYIVIPS 157
V DD++++P+
Sbjct 390 VNDDFVLLPA 399
Lambda K H
0.321 0.138 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3407623970
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40