bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0333_orf1
Length=176
Score E
Sequences producing significant alignments: (Bits) Value
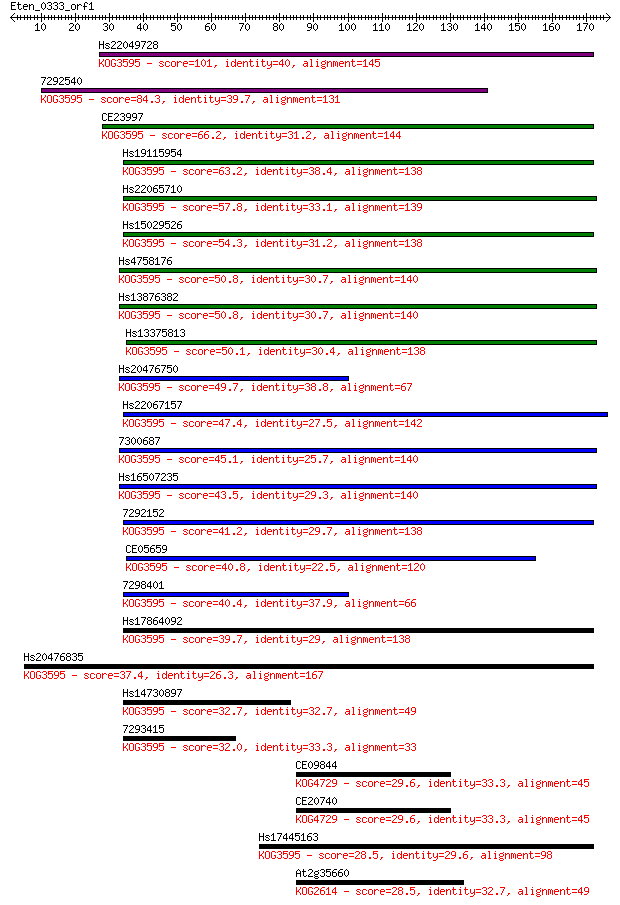
Hs22049728 101 8e-22
7292540 84.3 1e-16
CE23997 66.2 3e-11
Hs19115954 63.2 2e-10
Hs22065710 57.8 1e-08
Hs15029526 54.3 1e-07
Hs4758176 50.8 1e-06
Hs13876382 50.8 2e-06
Hs13375813 50.1 3e-06
Hs20476750 49.7 3e-06
Hs22067157 47.4 2e-05
7300687 45.1 7e-05
Hs16507235 43.5 2e-04
7292152 41.2 0.001
CE05659 40.8 0.001
7298401 40.4 0.002
Hs17864092 39.7 0.003
Hs20476835 37.4 0.015
Hs14730897 32.7 0.35
7293415 32.0 0.62
CE09844 29.6 3.0
CE20740 29.6 3.2
Hs17445163 28.5 7.3
At2g35660 28.5 7.7
> Hs22049728
Length=2274
Score = 101 bits (251), Expect = 8e-22, Method: Compositional matrix adjust.
Identities = 58/147 (39%), Positives = 87/147 (59%), Gaps = 8/147 (5%)
Query 27 ELSQWRVWLGGLFFPSAYLTATRQAVAQAKGWSLDDLAVEVLVGSVEAK--DDQSFIITG 84
EL V LGGLF P AY+TATRQ VAQA WSL++L +EV V + + D SF +TG
Sbjct 2131 ELKNIHVCLGGLFVPEAYITATRQYVAQANSWSLEELCLEVNVTTSQGATLDACSFGVTG 2190
Query 85 LTVEGAAWNANEKCLDVADTIASSLPPMVMRWYHKGEGCKFKEDGQTYISTPLFLNRARK 144
L ++GA N N+ L +++ I+++LP +RW + E + ++ P++LN R
Sbjct 2191 LKLQGATCNNNK--LSLSNAISTALPLTQLRWVKQTNT----EKKASVVTLPVYLNFTRA 2244
Query 145 DLVAFVDLPASEQYPPPLWVQRGMAVL 171
DL+ VD + + P + +RG+AVL
Sbjct 2245 DLIFTVDFEIATKEDPRSFYERGVAVL 2271
> 7292540
Length=4680
Score = 84.3 bits (207), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 52/134 (38%), Positives = 76/134 (56%), Gaps = 11/134 (8%)
Query 10 QQIEAAMNLLSRQRDDGELSQWRVWLGGLFFPSAYLTATRQAVAQAKGWSLDDLAVEVLV 69
QQ++ L+S Q EL + VWLGGL P AY+TATRQ VAQA WSL++LA++V +
Sbjct 4461 QQLQKVSQLVS-QAGAKELQGFPVWLGGLLNPEAYITATRQCVAQANSWSLEELALDVTI 4519
Query 70 GSVEAKDDQS---FIITGLTVEGAAWNANEKCLDVADTIASSLPPMVMRWYHKGEGCKFK 126
K+DQ F +TGL ++GA NE L +A TI LP +++W +
Sbjct 4520 TDAGLKNDQKDCCFGVTGLKLQGAQCKNNE--LLLASTIMMDLPVTILKWIKISSEPRIS 4577
Query 127 EDGQTYISTPLFLN 140
+ ++ P++LN
Sbjct 4578 K-----LTLPVYLN 4586
> CE23997
Length=4568
Score = 66.2 bits (160), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 45/144 (31%), Positives = 72/144 (50%), Gaps = 15/144 (10%)
Query 28 LSQWRVWLGGLFFPSAYLTATRQAVAQAKGWSLDDLAVEVLVGSVEAKDDQSFIITGLTV 87
L + WLGG F P AY+TATRQ VAQA WSL+ L + + +G ++ D F I+G+ +
Sbjct 4435 LKRETFWLGGTFSPEAYITATRQQVAQANTWSLEQLNLHIHIGRTDSTD--VFRISGIDI 4492
Query 88 EGAAWNANEKCLDVADTIASSLPPMVMRWYHKGEGCKFKEDGQTYISTPLFLNRARKDLV 147
GA K L++ + + S + W K+D PL+L R+ L+
Sbjct 4493 RGAKSVGGNK-LELCELVKSECDIVEFSW---------KQDVADGTRLPLYLYGDRRQLI 4542
Query 148 AFVDLPASEQYPPPLWVQRGMAVL 171
+ + S ++ QRG+A++
Sbjct 4543 SPLAFHLS---SATVFYQRGVALV 4563
> Hs19115954
Length=4624
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 53/150 (35%), Positives = 77/150 (51%), Gaps = 22/150 (14%)
Query 34 WLGGLFFPSAYLTATRQAVAQA-KGWSLDDLAVEVLVGSVEA--KDDQS------FIITG 84
W+ G F P +LTA RQ + +A KGW+LD++ VL V KDD S + G
Sbjct 4481 WMTGFFNPQGFLTAMRQEITRANKGWALDNM---VLCNEVTKWMKDDISAPPTEGVYVYG 4537
Query 85 LTVEGAAWNA-NEKCLDVADTIASSLPPMVMRWYHKGEGCKFKEDGQTYISTPLFLNRAR 143
L +EGA W+ N K ++ + L P V+R Y + + D + Y S P++ R
Sbjct 4538 LYLEGAGWDKRNMKLIESKPKVLFELMP-VIRIYAENNTLR---DPRFY-SCPIYKKPVR 4592
Query 144 KDL--VAFVDLPASEQYPPPLWVQRGMAVL 171
DL +A VDL ++ P WV RG+A+L
Sbjct 4593 TDLNYIAAVDLRTAQ--TPEHWVLRGVALL 4620
> Hs22065710
Length=323
Score = 57.8 bits (138), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 46/152 (30%), Positives = 69/152 (45%), Gaps = 19/152 (12%)
Query 34 WLGGLFFPSAYLTATRQAVAQAKGWSLDDLAVEVLVGSVEAKDDQSFI--------ITGL 85
WL G FP+ +LTA Q+ A+ S+D L+ E +V +V DD + + + GL
Sbjct 174 WLSGFTFPTGFLTAVLQSSARQNNVSVDSLSWEFIVSTV---DDSNLVYPPKDGVWVRGL 230
Query 86 TVEGAAWNANEKCLDVADTIA-SSLPPMVMRWYHKGEGCKFKEDGQTYISTPLFLNRA-- 142
+EGA W+ CL A+ + L P + + E K G + NRA
Sbjct 231 YLEGAGWDRKNSCLVEAEPMQLVCLMPTIH--FRPAESRKKSAKGMYSCPCYYYPNRAGS 288
Query 143 --RKDLVAFVDLPASEQYPPPLWVQRGMAVLL 172
R V +DL S P W++RG A+L+
Sbjct 289 SDRASFVIGIDL-RSGAMTPDHWIKRGTALLM 319
> Hs15029526
Length=4490
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 43/145 (29%), Positives = 70/145 (48%), Gaps = 11/145 (7%)
Query 34 WLGGLFFPSAYLTATRQAVAQA-KGWSLDDLAV--EVLVGSVE---AKDDQSFIITGLTV 87
W+ G F P +LTA RQ V +A KGW+LD + + EVL + E + + I GL +
Sbjct 4346 WMTGFFNPQGFLTAMRQEVTRAHKGWALDTVTIHNEVLRQTKEEITSPPGEGVYIYGLYM 4405
Query 88 EGAAWN-ANEKCLDVADTIASSLPPMVMRWYHKGEGCKFKEDGQTYISTPLFLNRARKDL 146
+GAAW+ N K ++ + + P++ + K D + Y+ P++ R DL
Sbjct 4406 DGAAWDRRNGKLMESTPKVLFTQLPVLHIFAINSTAPK---DPKLYVC-PIYKKPRRTDL 4461
Query 147 VAFVDLPASEQYPPPLWVQRGMAVL 171
+ P W+ RG+A+L
Sbjct 4462 TFITVVYLRTVLSPDHWILRGVALL 4486
> Hs4758176
Length=798
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 43/149 (28%), Positives = 67/149 (44%), Gaps = 17/149 (11%)
Query 33 VWLGGLFFPSAYLTATRQAVAQAKGWSLDDLAVEVLVGSVEAKDDQSF--------IITG 84
VWL G F P ++LTA Q+ A+ W LD +A++ + K+ + F I G
Sbjct 656 VWLTGFFNPQSFLTAIMQSTARKNEWPLDQMALQC---DMTKKNREEFRSPPREGAYIHG 712
Query 85 LTVEGAAWNANEKCLDVADTIASSLPPMVMRWYHKGEGCKFKEDGQTYISTPLFLNRAR- 143
L +EGA W+ + A + PPM + + K+D ++ S P++ R
Sbjct 713 LFMEGACWDTQAGIITEAK-LKDLTPPMPVMFIKAIPAD--KQDCRSVYSCPVYKTSQRG 769
Query 144 KDLVAFVDLPASEQYPPPLWVQRGMAVLL 172
V +L E P WV G+A+LL
Sbjct 770 PTYVWTFNLKTKEN--PSKWVLAGVALLL 796
> Hs13876382
Length=4486
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 43/149 (28%), Positives = 67/149 (44%), Gaps = 17/149 (11%)
Query 33 VWLGGLFFPSAYLTATRQAVAQAKGWSLDDLAVEVLVGSVEAKDDQSF--------IITG 84
VWL G F P ++LTA Q+ A+ W LD +A++ + K+ + F I G
Sbjct 4344 VWLTGFFNPQSFLTAIMQSTARKNEWPLDQMALQC---DMTKKNREEFRSPPREGAYIHG 4400
Query 85 LTVEGAAWNANEKCLDVADTIASSLPPMVMRWYHKGEGCKFKEDGQTYISTPLFLNRAR- 143
L +EGA W+ + A + PPM + + K+D ++ S P++ R
Sbjct 4401 LFMEGACWDTQAGIITEAK-LKDLTPPMPVMFIKAIPAD--KQDCRSVYSCPVYKTSQRG 4457
Query 144 KDLVAFVDLPASEQYPPPLWVQRGMAVLL 172
V +L E P WV G+A+LL
Sbjct 4458 PTYVWTFNLKTKEN--PSKWVLAGVALLL 4484
> Hs13375813
Length=553
Score = 50.1 bits (118), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 42/140 (30%), Positives = 67/140 (47%), Gaps = 7/140 (5%)
Query 35 LGGLFFPSAYLTATRQAVAQAKGWSLDDLA-VEVLVGSVEAKDDQSFIITGLTVEGAAWN 93
L LF P +L A RQ A+A G S+D L V G ++ Q I+GL +EG +++
Sbjct 416 LSELFHPDTFLNALRQETARAVGRSVDSLKFVASWKGRLQEAKLQ-IKISGLLLEGCSFD 474
Query 94 ANEKCLDVADTIA-SSLPPMVMRWYHKGEGCKFKEDGQTYISTPLFLNRARKDLVAFVDL 152
N+ + D+ + SS+ P M W + + D IS P++ + R +V +D+
Sbjct 475 GNQLSENQLDSPSVSSVLPCFMGWIPQDARGPYSPD--ECISLPVYTSAERDRVVTNIDV 532
Query 153 PASEQYPPPLWVQRGMAVLL 172
P W+Q G A+ L
Sbjct 533 PCGGNQDQ--WIQCGAALFL 550
> Hs20476750
Length=223
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 26/73 (35%), Positives = 37/73 (50%), Gaps = 6/73 (8%)
Query 33 VWLGGLFFPSAYLTATRQAVAQAKGWSLDDLAVEVLVGSVEAKDD------QSFIITGLT 86
+WL GL P +YLTA QA + GW LD + V + D+ Q ++GL
Sbjct 75 MWLSGLHIPESYLTALVQATCRKNGWPLDRSTLFTQVTKFQDADEVNERAGQGCFVSGLY 134
Query 87 VEGAAWNANEKCL 99
+EGA W+ + CL
Sbjct 135 LEGADWDIEKGCL 147
> Hs22067157
Length=3672
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 39/157 (24%), Positives = 65/157 (41%), Gaps = 21/157 (13%)
Query 34 WLGGLFFPSAYLTATRQAVAQAKGWSLDDLAVEVLVGS----VEAKDDQSFIITGLTVEG 89
W+ G +F ++LT Q A+ +D + E V +E + I GL +EG
Sbjct 3521 WISGFYFTQSFLTGVSQNYARKYTIPIDHIGFEFEVTPQETVMENNPEDGAYIKGLFLEG 3580
Query 90 AAWNANEKCLDVADTIASSL-PPMVMRWYHKGEGCKFKEDGQTYISTPLFLNRARK---- 144
A W + K + + +++ L P+ + W GE F Q P++ AR+
Sbjct 3581 ARW--DRKTMQIGESLPKILYDPLPIIWLKPGESAMFLH--QDIYVCPVYKTSARRGTLS 3636
Query 145 ------DLVAFVDLPASEQYPPPLWVQRGMAVLLWGD 175
+ V ++LP P W+ RG+A L D
Sbjct 3637 TTGHSTNYVLSIELPTD--MPQKHWINRGVASLCQLD 3671
> 7300687
Length=4472
Score = 45.1 bits (105), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 36/150 (24%), Positives = 63/150 (42%), Gaps = 18/150 (12%)
Query 33 VWLGGLFFPSAYLTATRQAVAQAKGWSLDDLAVEVLVGSVEAKDDQSF--------IITG 84
VWL G F P + LTA Q+ A+ LD + ++ V K + F + G
Sbjct 4329 VWLAGFFNPQSLLTAIMQSTARRNDLPLDKMCLQC---DVTKKQKEEFTTAPRDGCCVHG 4385
Query 85 LTVEGAAWNANEKCL--DVADTIASSLPPMVMRWYHKGEGCKFKEDGQTYISTPLFLNRA 142
+ +EGA W+ + + + S+P + +R + K+D + P++ R
Sbjct 4386 IFMEGARWDIQQGIIMESRLKELYPSMPVINIRAITQD-----KQDLRNMYECPVYKTRT 4440
Query 143 RKDLVAFVDLPASEQYPPPLWVQRGMAVLL 172
R +L + P W+ G+A+LL
Sbjct 4441 RGPTTYVSNLNLKTKDKPGKWILAGVALLL 4470
> Hs16507235
Length=4523
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 41/148 (27%), Positives = 62/148 (41%), Gaps = 15/148 (10%)
Query 33 VWLGGLFFPSAYLTATRQAVAQAKGWSLDDLAVEVLVGSVEAKDDQSF--------IITG 84
VWL G F P ++LTA Q +A+ W LD L V K + + + G
Sbjct 4381 VWLSGFFNPQSFLTAIMQTMARKNEWPLDKTR---LTADVTKKTKEDYGHPPREGAYLHG 4437
Query 85 LTVEGAAWNANEKCLDVADTIASSLPPMVMRWYHKGEGCKFKEDGQTYISTPLFLNRARK 144
L +EGA W+ + A + P V+ + K +E QTY P++ + R
Sbjct 4438 LFMEGARWDTQAGTIVEARLKELACPMPVI--FAKATPVDRQETKQTY-ECPVYRTKLRG 4494
Query 145 DLVAFVDLPASEQYPPPLWVQRGMAVLL 172
+ SE+ WV G+A+LL
Sbjct 4495 PSYIWTFRLKSEEKTAK-WVLAGVALLL 4521
> 7292152
Length=3868
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 41/156 (26%), Positives = 60/156 (38%), Gaps = 28/156 (17%)
Query 34 WLGGLFFPSAYLTATRQAVAQAKGWSLDDLAVEVLVGSVEAKD---DQSFIITGLTVEGA 90
WL G FF A+LT Q A+ +D L + V VE K D GL +EGA
Sbjct 3718 WLSGFFFTQAFLTGAMQNFARKYKIPIDTLTFDYDVLKVETKTSPPDDGVYCNGLYLEGA 3777
Query 91 AWNANEKCL--DVADTIASSLPPMVMR---WYHKGEGCKFKEDGQTYISTPLFLNRARKD 145
W E L + ++P + R EG +++ PL+ RK
Sbjct 3778 RWEWRENTLVEQFPKVLIYAMPVIFFRPVGLVDVVEGSRYR--------CPLYKTAERKG 3829
Query 146 LVAFVDLPASEQYPPPL----------WVQRGMAVL 171
++ S Y PL WV+R +A++
Sbjct 3830 TLSTTG--HSTNYVVPLLLNTHVKASHWVKRSVALI 3863
> CE05659
Length=4131
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 27/120 (22%), Positives = 51/120 (42%), Gaps = 4/120 (3%)
Query 35 LGGLFFPSAYLTATRQAVAQAKGWSLDDLAVEVLVGSVEAKDDQSFIITGLTVEGAAWNA 94
LF+P+ +L A RQ ++ LD L + + Q + GL ++GA +++
Sbjct 4000 FSDLFYPNIFLNALRQTTSRQIKIPLDQLILSSAWTPSQLPAKQCVQVQGLLLQGATFDS 4059
Query 95 NEKCLDVADTIASSLPPMVMRWYHKGEGCKFKEDGQTYISTPLFLNRARKDLVAFVDLPA 154
+ V+ S P + + W + E I P++ + R DL+ V++P
Sbjct 4060 FLRETTVSSAAYSQAPIVFLAWTSESSSTITGEQ----IQVPVYSSSERSDLICSVNMPC 4115
> 7298401
Length=4010
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 35/73 (47%), Gaps = 7/73 (9%)
Query 34 WLGGLFFPSAYLTATRQAVAQAKGWSLDDLAVEVLVGSVEAKDDQSF-------IITGLT 86
WL G FF A+LT +Q A+ S+D LA + V +VE Q + G+
Sbjct 3854 WLSGFFFTQAFLTGAQQNYARKYVISIDLLAFDYEVLTVEEPQRQGLSGPEDGVFVYGIF 3913
Query 87 VEGAAWNANEKCL 99
+EGA W+ K L
Sbjct 3914 LEGARWDRTGKYL 3926
> Hs17864092
Length=4024
Score = 39.7 bits (91), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 40/155 (25%), Positives = 71/155 (45%), Gaps = 26/155 (16%)
Query 34 WLGGLFFPSAYLTATRQAVAQAKGWSLDDLAVEVLVGSVEAKD-----DQSFIITGLTVE 88
WL G FF A+LT +Q A+ +D L + V +E K+ + I GL ++
Sbjct 3874 WLSGFFFTQAFLTGAQQNYARKYTIPIDLLGFDYEV--MEDKEYKHPPEDGVFIHGLFLD 3931
Query 89 GAAWNANEKCLDVAD--TIASSLPPMVMRWYHKGEGCKFKEDGQTYISTPLFLNRARK-- 144
GA+WN K L + + ++P M ++ + + K +Y++ PL+ R+
Sbjct 3932 GASWNRKIKKLAESHPKILYDTVPVMWLKPCKRADIPK----RPSYVA-PLYKTSERRGV 3986
Query 145 --------DLVAFVDLPASEQYPPPLWVQRGMAVL 171
+ V + LP+ + P W+ RG+A+L
Sbjct 3987 LSTTGHSTNFVIAMTLPSDQ--PKEHWIGRGVALL 4019
> Hs20476835
Length=209
Score = 37.4 bits (85), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 44/181 (24%), Positives = 72/181 (39%), Gaps = 21/181 (11%)
Query 5 PLGND-QQIEAAMNLLSRQRDDGELSQWRVWLGGLFFPSAYLTATRQAVAQAKGWSLDDL 63
PLG+ A +N L + G+ + WL G FF A+LT Q A+ +D L
Sbjct 31 PLGSYITDFLARLNFLQDWYNSGKPCVF--WLSGFFFTQAFLTGAMQNYARKYTTPIDLL 88
Query 64 AVEVLVGSVEAKD---DQSFIITGLTVEGAAWNANEKCL-DVADTIASSLPPMVMRWYHK 119
E V + D + I GL ++GA W+ L + + L P++ W
Sbjct 89 GYEFEVIPSDTSDTSPEDGVYIHGLYLDGARWDRESGLLAEQYPKLLFDLMPII--WIKP 146
Query 120 GEGCK-FKEDGQTYISTPLFLNRARKDLVA--------FVDLPASEQYPPPLWVQRGMAV 170
+ + K D PL+ RK ++ + + P W++RG+A+
Sbjct 147 TQKSRIIKSDAYV---CPLYKTSERKGTLSTTGHSTNFVIAMLLKTDQPTRHWIKRGVAL 203
Query 171 L 171
L
Sbjct 204 L 204
> Hs14730897
Length=1350
Score = 32.7 bits (73), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 26/49 (53%), Gaps = 2/49 (4%)
Query 34 WLGGLFFPSAYLTATRQAVAQAKGWSLDDLAVEVLVGSVEAKDDQSFII 82
W+ G FFP +LT T Q A+ +D+L+ + V + DQ+ +I
Sbjct 1179 WISGFFFPQGFLTGTLQNHARKYNLPIDELSFKYSV--IPTYRDQAAVI 1225
> 7293415
Length=4081
Score = 32.0 bits (71), Expect = 0.62, Method: Compositional matrix adjust.
Identities = 11/33 (33%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 34 WLGGLFFPSAYLTATRQAVAQAKGWSLDDLAVE 66
W+ G FFP ++LT Q A+ + +D L ++
Sbjct 3909 WISGFFFPQSFLTGVLQTYARRRVLPIDSLKID 3941
> CE09844
Length=460
Score = 29.6 bits (65), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 21/48 (43%), Gaps = 3/48 (6%)
Query 85 LTVEGAAWNANEKC---LDVADTIASSLPPMVMRWYHKGEGCKFKEDG 129
L V A + +C ++ S + P V+R H EGC K DG
Sbjct 188 LAVYSAQMKTSPQCDPETEIRHECVSDVLPQVLRQCHAKEGCTLKSDG 235
> CE20740
Length=464
Score = 29.6 bits (65), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 21/48 (43%), Gaps = 3/48 (6%)
Query 85 LTVEGAAWNANEKC---LDVADTIASSLPPMVMRWYHKGEGCKFKEDG 129
L V A + +C ++ S + P V+R H EGC K DG
Sbjct 192 LAVYSAQMKTSPQCDPETEIRHECVSDVLPQVLRQCHAKEGCTLKSDG 239
> Hs17445163
Length=133
Score = 28.5 bits (62), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 29/117 (24%), Positives = 42/117 (35%), Gaps = 24/117 (20%)
Query 74 AKDDQSFIITGLTVEGAAWNANEK----------CLDVADTIASSLPPMVMRWYHKGEGC 123
A I GL +EGA WN +K C D D LP + K
Sbjct 16 ASSHTGVYIFGLFIEGARWNREQKILEDSLPLEMCCDFPDIYF--LPTKIST---KTPNA 70
Query 124 KFKEDGQTY-ISTPLFLNRARKDLVAFVDLPA--------SEQYPPPLWVQRGMAVL 171
+ D + Y P++ R ++A LP S + PP W+ +A+L
Sbjct 71 SNQTDSELYAFECPVYQTPERSRILATTGLPTNFLTSVYLSTKKPPSHWITMRVALL 127
> At2g35660
Length=412
Score = 28.5 bits (62), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 27/49 (55%), Gaps = 1/49 (2%)
Query 85 LTVEGAAWNANEKCLDVADTIASSLPPMVMRWYHKGEGCKFKEDGQTYI 133
L +EG A E+ + + +T+AS LPP +R+ K E + +G T +
Sbjct 104 LEIEGQEVRAVERRV-LLETLASQLPPQTIRFSSKLESIQSNANGDTLL 151
Lambda K H
0.319 0.136 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2707167450
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40