bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0337_orf2
Length=109
Score E
Sequences producing significant alignments: (Bits) Value
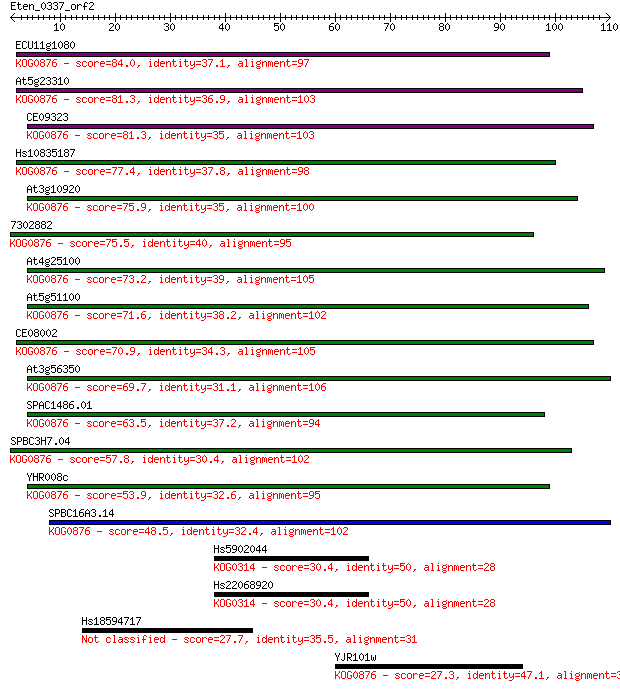
ECU11g1080 84.0 7e-17
At5g23310 81.3 4e-16
CE09323 81.3 5e-16
Hs10835187 77.4 6e-15
At3g10920 75.9 2e-14
7302882 75.5 3e-14
At4g25100 73.2 1e-13
At5g51100 71.6 3e-13
CE08002 70.9 6e-13
At3g56350 69.7 1e-12
SPAC1486.01 63.5 9e-11
SPBC3H7.04 57.8 6e-09
YHR008c 53.9 8e-08
SPBC16A3.14 48.5 3e-06
Hs5902044 30.4 0.90
Hs22068920 30.4 0.90
Hs18594717 27.7 4.9
YJR101w 27.3 6.8
> ECU11g1080
Length=233
Score = 84.0 bits (206), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 36/97 (37%), Positives = 54/97 (55%), Gaps = 0/97 (0%)
Query 2 EELESAFGGVENFREAFAAAAAAHFGSGWAWLCFCKKSRSLFLLQTHDGATPFRDNPNCA 61
E +E +FG E + F+ AA + FGSGW WLC+ + L + +T++ +
Sbjct 113 EYIERSFGSQEKMVKEFSNAAVSLFGSGWVWLCYRQSEGILVIRKTYNQDAICMKTSSTV 172
Query 62 PLLTCDLWEHAYYIDRRNDRKSYLDAWWSVVNWDFAN 98
P+L D+WEHAYY+ +N R Y+ WW VVNW +
Sbjct 173 PILGLDVWEHAYYLKYKNVRSEYVANWWKVVNWGLVS 209
> At5g23310
Length=263
Score = 81.3 bits (199), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 38/105 (36%), Positives = 63/105 (60%), Gaps = 5/105 (4%)
Query 2 EELESAFGGVENFREAFAAAAAAHFGSGWAWLCFCKKSRSLFLLQTHDGATPFR-DNPNC 60
E+++ FG NFRE F AA FGSGW WL ++ R L +++T + P D+
Sbjct 148 EQIDKDFGSFTNFREKFTNAALTQFGSGWVWLVLKREERRLEVVKTSNAINPLVWDD--- 204
Query 61 APLLTCDLWEHAYYIDRRNDRKSYLDAWWS-VVNWDFANENLKKA 104
P++ D+WEH+YY+D +NDR Y++ + + +V+W+ A + +A
Sbjct 205 IPIICVDVWEHSYYLDYKNDRAKYINTFLNHLVSWNAAMSRMARA 249
> CE09323
Length=221
Score = 81.3 bits (199), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 36/103 (34%), Positives = 55/103 (53%), Gaps = 1/103 (0%)
Query 4 LESAFGGVENFREAFAAAAAAHFGSGWAWLCFCKKSRSLFLLQTHDGATPFRDNPNCAPL 63
++S FG ++N ++ +A+ A GSGW WL +C K + + + T P PL
Sbjct 120 IKSDFGSLDNLQKQLSASTVAVQGSGWGWLGYCPKGK-ILKVATCANQDPLEATTGLVPL 178
Query 64 LTCDLWEHAYYIDRRNDRKSYLDAWWSVVNWDFANENLKKAMQ 106
D+WEHAYY+ +N R Y++A W + NW +E KA Q
Sbjct 179 FGIDVWEHAYYLQYKNVRPDYVNAIWKIANWKNVSERFAKAQQ 221
> Hs10835187
Length=222
Score = 77.4 bits (189), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 37/98 (37%), Positives = 50/98 (51%), Gaps = 1/98 (1%)
Query 2 EELESAFGGVENFREAFAAAAAAHFGSGWAWLCFCKKSRSLFLLQTHDGATPFRDNPNCA 61
E ++ FG + F+E AA+ GSGW WL F K R + P +
Sbjct 119 EAIKLDFGSFDKFKEKLTAASVGVQGSGWGWLGF-NKERGHLQIAACPNQDPLQGTTGLI 177
Query 62 PLLTCDLWEHAYYIDRRNDRKSYLDAWWSVVNWDFANE 99
PLL D+WEHAYY+ +N R YL A W+V+NW+ E
Sbjct 178 PLLGIDVWEHAYYLQYKNVRPDYLKAIWNVINWENVTE 215
> At3g10920
Length=231
Score = 75.9 bits (185), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 35/100 (35%), Positives = 54/100 (54%), Gaps = 0/100 (0%)
Query 4 LESAFGGVENFREAFAAAAAAHFGSGWAWLCFCKKSRSLFLLQTHDGATPFRDNPNCAPL 63
+++ FG +E + +A AA GSGW WL K+ + L + T + + PL
Sbjct 129 IDAHFGSLEGLVKKMSAEGAAVQGSGWVWLGLDKELKKLVVDTTANQDPLVTKGGSLVPL 188
Query 64 LTCDLWEHAYYIDRRNDRKSYLDAWWSVVNWDFANENLKK 103
+ D+WEHAYY+ +N R YL W V+NW +A+E +K
Sbjct 189 VGIDVWEHAYYLQYKNVRPEYLKNVWKVINWKYASEVYEK 228
> 7302882
Length=217
Score = 75.5 bits (184), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 38/97 (39%), Positives = 52/97 (53%), Gaps = 5/97 (5%)
Query 1 REELESAFGGVENFREAFAAAAAAHFGSGWAWLCFCKKSRSLFL--LQTHDGATPFRDNP 58
++ +ES + +E F++ A GSGW WL F KKS L L L D P +
Sbjct 110 KKAIESQWKSLEEFKKELTTLTVAVQGSGWGWLGFNKKSGKLQLAALPNQD---PLEAST 166
Query 59 NCAPLLTCDLWEHAYYIDRRNDRKSYLDAWWSVVNWD 95
PL D+WEHAYY+ +N R SY++A W + NWD
Sbjct 167 GLIPLFGIDVWEHAYYLQYKNVRPSYVEAIWDIANWD 203
> At4g25100
Length=212
Score = 73.2 bits (178), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 41/106 (38%), Positives = 59/106 (55%), Gaps = 5/106 (4%)
Query 4 LESAFGGVENFREAFAAAAAAHFGSGWAWLCFCKKSRSLFLLQTHDGATPFRDNPNCAPL 63
LE F E F E F AAAA FG+GWAWL + + L +++T + P PL
Sbjct 110 LERDFTSYEKFYEEFNAAAATQFGAGWAWLAYS--NEKLKVVKTPNAVNPLVL--GSFPL 165
Query 64 LTCDLWEHAYYIDRRNDRKSYLDAWWS-VVNWDFANENLKKAMQGS 108
LT D+WEHAYY+D +N R Y+ + + +V+W+ + L+ A S
Sbjct 166 LTIDVWEHAYYLDFQNRRPDYIKTFMTNLVSWEAVSARLEAAKAAS 211
> At5g51100
Length=305
Score = 71.6 bits (174), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 39/118 (33%), Positives = 63/118 (53%), Gaps = 18/118 (15%)
Query 4 LESAFGGVENFREAFAAAAAAHFGSGWAWLCF---------------CKKSRSLFLLQTH 48
+E FG E F E F +AAA++FGSGW WL + ++ + L +++T
Sbjct 152 IERDFGSFEEFLERFKSAAASNFGSGWTWLAYKANRLDVANAVNPLPKEEDKKLVIVKTP 211
Query 49 DGATPFRDNPNCAPLLTCDLWEHAYYIDRRNDRKSYLDAWW-SVVNWDFANENLKKAM 105
+ P + +PLLT D WEHAYY+D N R Y++ + +V+W+ + L+ A+
Sbjct 212 NAVNPLVW--DYSPLLTIDTWEHAYYLDFENRRAEYINTFMEKLVSWETVSTRLESAI 267
> CE08002
Length=218
Score = 70.9 bits (172), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 36/107 (33%), Positives = 54/107 (50%), Gaps = 8/107 (7%)
Query 2 EELESAFGGVENFREAFAAAAAAHFGSGWAWLCFCKKSRSLFLLQTHDGATPFRDNP--N 59
+ ++ FG ++N ++ + A GSGW WL +CKK + L + AT +P
Sbjct 118 DTIKRDFGSLDNLQKRLSDITIAVQGSGWGWLGYCKKDKILKI------ATCANQDPLEG 171
Query 60 CAPLLTCDLWEHAYYIDRRNDRKSYLDAWWSVVNWDFANENLKKAMQ 106
PL D+WEHAYY+ +N R Y+ A W + NW +E A Q
Sbjct 172 MVPLFGIDVWEHAYYLQYKNVRPDYVHAIWKIANWKNISERFANARQ 218
> At3g56350
Length=241
Score = 69.7 bits (169), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 33/106 (31%), Positives = 53/106 (50%), Gaps = 0/106 (0%)
Query 4 LESAFGGVENFREAFAAAAAAHFGSGWAWLCFCKKSRSLFLLQTHDGATPFRDNPNCAPL 63
+++ FG +E + A AA GSGW W ++ + L + T + + PL
Sbjct 134 IDAHFGSLEGLIQKMNAEGAAVQGSGWVWFGLDRELKRLVVETTANQDPLVTKGSHLVPL 193
Query 64 LTCDLWEHAYYIDRRNDRKSYLDAWWSVVNWDFANENLKKAMQGSD 109
+ D+WEHAYY +N R YL W+V+NW +A + +K + D
Sbjct 194 IGIDVWEHAYYPQYKNARAEYLKNIWTVINWKYAADVFEKHTRDLD 239
> SPAC1486.01
Length=218
Score = 63.5 bits (153), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 35/94 (37%), Positives = 50/94 (53%), Gaps = 5/94 (5%)
Query 4 LESAFGGVENFREAFAAAAAAHFGSGWAWLCFCKKSRSLFLLQTHDGATPFRDNPNCAPL 63
+ S +G +E+F++ AA A+ GSGWAWL K SL + T + T + P+
Sbjct 123 ITSKWGSLEDFQKEMNAALASIQGSGWAWL-IVDKDGSLRITTTANQDTIVK----SKPI 177
Query 64 LTCDLWEHAYYIDRRNDRKSYLDAWWSVVNWDFA 97
+ D WEHAYY N + Y A W+V+NW A
Sbjct 178 IGIDAWEHAYYPQYENRKAEYFKAIWNVINWKEA 211
> SPBC3H7.04
Length=220
Score = 57.8 bits (138), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 31/112 (27%), Positives = 50/112 (44%), Gaps = 11/112 (9%)
Query 1 REELESAFGGVENFREAFAAAAAAHFGSGWAWLCFCKKSRSLFLLQTHD---------GA 51
+ ++++FG + + FG GW WL + + LL T++ G
Sbjct 103 KPGIDASFGSFGELKSQMVDVGNSVFGDGWLWLVYSPEKSLFSLLCTYNASNAFLWGTGF 162
Query 52 TPFRDNPNCAPLLTCDLWEHAYYIDR-RNDRKSYLDAWWSVVNWDFANENLK 102
FR N PLL +LW++AY D N +K Y+ WW ++NW N +
Sbjct 163 PKFRTNA-IVPLLCVNLWQYAYLDDYGLNGKKMYITKWWDMINWTVVNNRFQ 213
> YHR008c
Length=233
Score = 53.9 bits (128), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 31/96 (32%), Positives = 48/96 (50%), Gaps = 4/96 (4%)
Query 4 LESAFGGVENFREAFAAAAAAHFGSGWAWLCF-CKKSRSLFLLQTHDGATPFRDNPNCAP 62
++ FG ++ + A GSGWA++ L ++QT++ T P P
Sbjct 133 IDEQFGSLDELIKLTNTKLAGVQGSGWAFIVKNLSNGGKLDVVQTYNQDT--VTGP-LVP 189
Query 63 LLTCDLWEHAYYIDRRNDRKSYLDAWWSVVNWDFAN 98
L+ D WEHAYY+ +N + Y A W+VVNW A+
Sbjct 190 LVAIDAWEHAYYLQYQNKKADYFKAIWNVVNWKEAS 225
> SPBC16A3.14
Length=277
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 33/125 (26%), Positives = 52/125 (41%), Gaps = 24/125 (19%)
Query 8 FGGVENFREAFAAAAAAHFGSGWAWLCFCKKSRSLFLLQTHDGATPF------RDNPNC- 60
FG EN A+ FG+ W W+ +R L LL+T +P+ ++P+
Sbjct 148 FGSKENLLSKIHELASNSFGACWLWIVIDDYNR-LNLLRTFQAGSPYLWTRWQSNDPHLI 206
Query 61 ---------------APLLTCDLWEHAYYIDR-RNDRKSYLDAWWSVVNWDFANENLKKA 104
P+L LW HAYY D +R Y+D W+ ++W E L +
Sbjct 207 SSVPDYSARPRKYAHVPILNLCLWNHAYYKDYGLLNRSRYIDTWFDCIDWSVIEERLTNS 266
Query 105 MQGSD 109
+ S+
Sbjct 267 LANSE 271
> Hs5902044
Length=948
Score = 30.4 bits (67), Expect = 0.90, Method: Composition-based stats.
Identities = 14/28 (50%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 38 KSRSLFLLQTHDGATPFRDNPNCAPLLT 65
K+ SLF+L + D ATP RD P A +T
Sbjct 794 KTDSLFVLPSRDDATPVRDEPMDAESIT 821
> Hs22068920
Length=1663
Score = 30.4 bits (67), Expect = 0.90, Method: Composition-based stats.
Identities = 14/28 (50%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 38 KSRSLFLLQTHDGATPFRDNPNCAPLLT 65
K+ SLF+L + D ATP RD P A +T
Sbjct 841 KTDSLFVLPSRDDATPVRDEPMDAESIT 868
> Hs18594717
Length=105
Score = 27.7 bits (60), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 11/31 (35%), Positives = 16/31 (51%), Gaps = 0/31 (0%)
Query 14 FREAFAAAAAAHFGSGWAWLCFCKKSRSLFL 44
+ F A +A+ GS W W C C S +L +
Sbjct 62 LKSGFRARSASLPGSQWTWFCMCSLSLTLAM 92
> YJR101w
Length=266
Score = 27.3 bits (59), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 16/40 (40%), Positives = 21/40 (52%), Gaps = 6/40 (15%)
Query 60 CAPLLTCDLWEHAYYID-----RRNDRKSYLDAW-WSVVN 93
P++ +LW+HAY D R K+ LD WSVVN
Sbjct 218 TMPIICVNLWDHAYLHDYGVGNRSKYVKNVLDNLNWSVVN 257
Lambda K H
0.322 0.134 0.464
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1199474506
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40