bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0339_orf4
Length=208
Score E
Sequences producing significant alignments: (Bits) Value
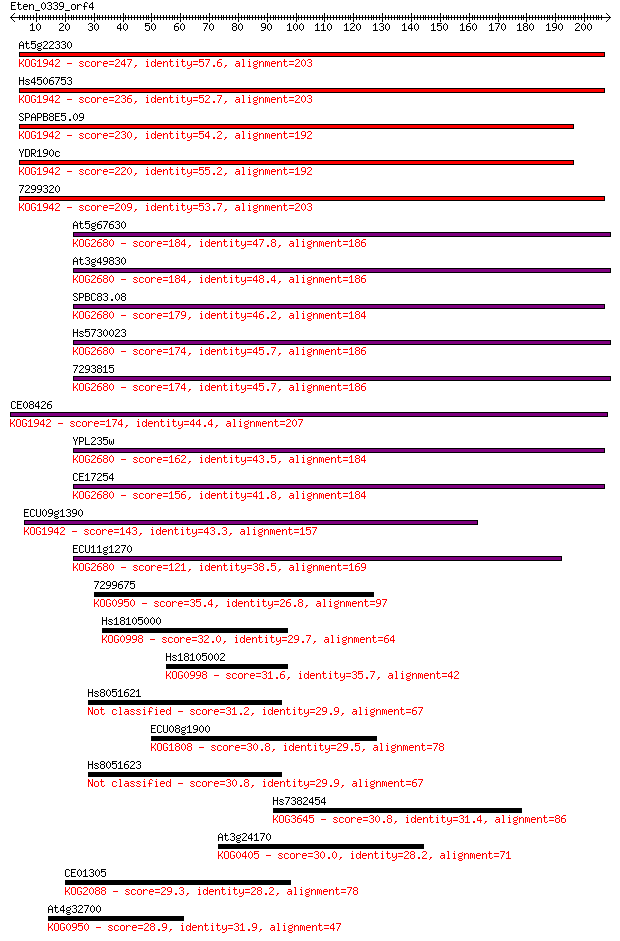
At5g22330 247 1e-65
Hs4506753 236 2e-62
SPAPB8E5.09 230 2e-60
YDR190c 220 1e-57
7299320 209 3e-54
At5g67630 184 9e-47
At3g49830 184 2e-46
SPBC83.08 179 5e-45
Hs5730023 174 9e-44
7293815 174 1e-43
CE08426 174 1e-43
YPL235w 162 5e-40
CE17254 156 3e-38
ECU09g1390 143 2e-34
ECU11g1270 121 1e-27
7299675 35.4 0.086
Hs18105000 32.0 0.85
Hs18105002 31.6 1.3
Hs8051621 31.2 1.7
ECU08g1900 30.8 1.8
Hs8051623 30.8 1.8
Hs7382454 30.8 2.2
At3g24170 30.0 3.8
CE01305 29.3 5.9
At4g32700 28.9 7.2
> At5g22330
Length=458
Score = 247 bits (631), Expect = 1e-65, Method: Compositional matrix adjust.
Identities = 117/203 (57%), Positives = 154/203 (75%), Gaps = 0/203 (0%)
Query 4 QVGTDVLSMMGAYMKPRKTEITERLREEINKTVNKYIEQGVAQLIPGVLFIDEVHMLDIE 63
Q G D+LS+MG MKPRKTEIT++LR+EINK VN+YI++GVA+L+PGVLFIDEVHMLD+E
Sbjct 254 QGGQDILSLMGQMMKPRKTEITDKLRQEINKVVNRYIDEGVAELVPGVLFIDEVHMLDME 313
Query 64 CFSFLNRVLESPMSPIIVFATNRGVSTVRGTDSVEPPGMPVDFLDRLLIIKTQPYTVSEV 123
CFS+LNR LES +SPI++FATNRGV VRGTD P G+P+D LDRL+II+TQ Y SE+
Sbjct 314 CFSYLNRALESSLSPIVIFATNRGVCNVRGTDMPSPHGVPIDLLDRLVIIRTQIYDPSEM 373
Query 124 VQVIQLRAAVERVNLSPSALEALGEIGGQISVRFALQLLEPCRLAAEARGSEVVEPNDVA 183
+Q+I +RA VE + + L LGEIG + S+R A+QLL P + A+ G + + D+
Sbjct 374 IQIIAIRAQVEELTVDEECLVLLGEIGQRTSLRHAVQLLSPASIVAKMNGRDNICKADIE 433
Query 184 DVDSLYLDAKASPVRLAEQPHRF 206
+V SLYLDAK+S L EQ ++
Sbjct 434 EVTSLYLDAKSSAKLLHEQQEKY 456
> Hs4506753
Length=456
Score = 236 bits (602), Expect = 2e-62, Method: Compositional matrix adjust.
Identities = 107/204 (52%), Positives = 154/204 (75%), Gaps = 1/204 (0%)
Query 4 QVGTDVLSMMGAYMKPRKTEITERLREEINKTVNKYIEQGVAQLIPGVLFIDEVHMLDIE 63
Q G D+LSMMG MKP+KTEIT++LR EINK VNKYI+QG+A+L+PGVLF+DEVHMLDIE
Sbjct 251 QGGQDILSMMGQLMKPKKTEITDKLRGEINKVVNKYIDQGIAELVPGVLFVDEVHMLDIE 310
Query 64 CFSFLNRVLESPMSPIIVFATNRGVSTVRGTDSVEPP-GMPVDFLDRLLIIKTQPYTVSE 122
CF++L+R LES ++PI++FA+NRG +RGT+ + P G+P+D LDR++II+T YT E
Sbjct 311 CFTYLHRALESSIAPIVIFASNRGNCVIRGTEDITSPHGIPLDLLDRVMIIRTMLYTPQE 370
Query 123 VVQVIQLRAAVERVNLSPSALEALGEIGGQISVRFALQLLEPCRLAAEARGSEVVEPNDV 182
+ Q+I++RA E +N+S AL LGEIG + ++R+++QLL P L A+ G + +E V
Sbjct 371 MKQIIKIRAQTEGINISEEALNHLGEIGTKTTLRYSVQLLTPANLLAKINGKDSIEKEHV 430
Query 183 ADVDSLYLDAKASPVRLAEQPHRF 206
++ L+ DAK+S LA+Q ++
Sbjct 431 EEISELFYDAKSSAKILADQQDKY 454
> SPAPB8E5.09
Length=456
Score = 230 bits (586), Expect = 2e-60, Method: Compositional matrix adjust.
Identities = 104/193 (53%), Positives = 148/193 (76%), Gaps = 1/193 (0%)
Query 4 QVGTDVLSMMGAYMKPRKTEITERLREEINKTVNKYIEQGVAQLIPGVLFIDEVHMLDIE 63
Q G D++SMMG MKP+KTEIT++LR EINK VNKYIEQG+A+LIPGVLFIDEVHMLDIE
Sbjct 252 QGGQDIMSMMGQLMKPKKTEITDKLRGEINKVVNKYIEQGIAELIPGVLFIDEVHMLDIE 311
Query 64 CFSFLNRVLESPMSPIIVFATNRGVSTVRGTDSVEPP-GMPVDFLDRLLIIKTQPYTVSE 122
CF++LN+ LES +SPI++FA+NRG+ T+RGT+ ++ P G+P D LDRLLI++T PY+ SE
Sbjct 312 CFTYLNQALESTISPIVIFASNRGICTIRGTEDIQAPHGIPTDLLDRLLIVRTLPYSESE 371
Query 123 VVQVIQLRAAVERVNLSPSALEALGEIGGQISVRFALQLLEPCRLAAEARGSEVVEPNDV 182
+ ++Q+RA VE + L+ L+ L + G + S+R+ +QLL P + A G++ + D+
Sbjct 372 IRSILQIRAKVENIILTDECLDKLAQEGSRTSLRYVIQLLTPVSIIASLHGNKEIGVQDI 431
Query 183 ADVDSLYLDAKAS 195
+ + L+LDA+ S
Sbjct 432 EECNDLFLDARRS 444
> YDR190c
Length=463
Score = 220 bits (561), Expect = 1e-57, Method: Compositional matrix adjust.
Identities = 106/193 (54%), Positives = 146/193 (75%), Gaps = 1/193 (0%)
Query 4 QVGTDVLSMMGAYMKPRKTEITERLREEINKTVNKYIEQGVAQLIPGVLFIDEVHMLDIE 63
Q G DV+SMMG +KP+KTEITE+LR+E+NK V KYI+QGVA+LIPGVLFIDEV+MLDIE
Sbjct 260 QGGQDVISMMGQLLKPKKTEITEKLRQEVNKVVAKYIDQGVAELIPGVLFIDEVNMLDIE 319
Query 64 CFSFLNRVLESPMSPIIVFATNRGVSTVRGT-DSVEPPGMPVDFLDRLLIIKTQPYTVSE 122
F++LN+ LES ++P++V A+NRG++TVRGT D + P G+P D +DRLLI++T PY E
Sbjct 320 IFTYLNKALESNIAPVVVLASNRGMTTVRGTEDVISPHGVPPDLIDRLLIVRTLPYDKDE 379
Query 123 VVQVIQLRAAVERVNLSPSALEALGEIGGQISVRFALQLLEPCRLAAEARGSEVVEPNDV 182
+ +I+ RA VER+ + SAL+ L +G + S+R+ALQLL PC + A+ + + NDV
Sbjct 380 IRTIIERRATVERLQVESSALDLLATMGTETSLRYALQLLAPCGILAQTSNRKEIVVNDV 439
Query 183 ADVDSLYLDAKAS 195
+ L+LDAK S
Sbjct 440 NEAKLLFLDAKRS 452
> 7299320
Length=456
Score = 209 bits (533), Expect = 3e-54, Method: Compositional matrix adjust.
Identities = 109/204 (53%), Positives = 151/204 (74%), Gaps = 1/204 (0%)
Query 4 QVGTDVLSMMGAYMKPRKTEITERLREEINKTVNKYIEQGVAQLIPGVLFIDEVHMLDIE 63
Q G DVLSMMG MKP+KTEIT++LR EINK VNKYI+QG+A+L+PGVLFIDE+HMLD+E
Sbjct 251 QGGQDVLSMMGQLMKPKKTEITDKLRMEINKVVNKYIDQGIAELVPGVLFIDEIHMLDLE 310
Query 64 CFSFLNRVLESPMSPIIVFATNRGVSTVRG-TDSVEPPGMPVDFLDRLLIIKTQPYTVSE 122
F++L++ LESP++PI++FATNRG +RG TD V P G+P+D LDRLLII+T Y+ ++
Sbjct 311 TFTYLHKSLESPIAPIVIFATNRGRCVIRGTTDIVSPHGIPLDLLDRLLIIRTLLYSTAD 370
Query 123 VVQVIQLRAAVERVNLSPSALEALGEIGGQISVRFALQLLEPCRLAAEARGSEVVEPNDV 182
+ Q+I+LRA E + L +A L EIG ++R+A+QLL P + G + +D+
Sbjct 371 MEQIIKLRAQTEGLQLEENAFTRLSEIGTSSTLRYAVQLLTPAHQMCKVNGRNQISKDDI 430
Query 183 ADVDSLYLDAKASPVRLAEQPHRF 206
DV SL+LDAK S L+E+ ++F
Sbjct 431 EDVHSLFLDAKRSSKHLSEKNNKF 454
> At5g67630
Length=469
Score = 184 bits (468), Expect = 9e-47, Method: Compositional matrix adjust.
Identities = 89/186 (47%), Positives = 127/186 (68%), Gaps = 0/186 (0%)
Query 23 EITERLREEINKTVNKYIEQGVAQLIPGVLFIDEVHMLDIECFSFLNRVLESPMSPIIVF 82
EI +RE+I+ V ++ E+G A+++PGVLFIDEVHMLDIECFSFLNR LE+ MSPI+V
Sbjct 264 EIRSEVREQIDTKVAEWREEGKAEIVPGVLFIDEVHMLDIECFSFLNRALENEMSPILVV 323
Query 83 ATNRGVSTVRGTDSVEPPGMPVDFLDRLLIIKTQPYTVSEVVQVIQLRAAVERVNLSPSA 142
ATNRGV+T+RGT+ P G+P+D LDRLLII TQPYT ++ +++++R E V ++ A
Sbjct 324 ATNRGVTTIRGTNQKSPHGIPIDLLDRLLIITTQPYTDDDIRKILEIRCQEEDVEMNEEA 383
Query 143 LEALGEIGGQISVRFALQLLEPCRLAAEARGSEVVEPNDVADVDSLYLDAKASPVRLAEQ 202
+ L IG S+R+A+ L+ L+ + R +VVE D+ V L+LD + S L E
Sbjct 384 KQLLTLIGRDTSLRYAIHLITAAALSCQKRKGKVVEVEDIQRVYRLFLDVRRSMQYLVEY 443
Query 203 PHRFPF 208
++ F
Sbjct 444 QSQYMF 449
> At3g49830
Length=473
Score = 184 bits (466), Expect = 2e-46, Method: Compositional matrix adjust.
Identities = 90/186 (48%), Positives = 123/186 (66%), Gaps = 0/186 (0%)
Query 23 EITERLREEINKTVNKYIEQGVAQLIPGVLFIDEVHMLDIECFSFLNRVLESPMSPIIVF 82
EI RE+ + V ++ E+G A+++PGVLFIDEVHMLDIECFSFLNR LE+ MSPI+V
Sbjct 265 EIRSETREQSDTKVAEWREEGKAEIVPGVLFIDEVHMLDIECFSFLNRALENDMSPILVV 324
Query 83 ATNRGVSTVRGTDSVEPPGMPVDFLDRLLIIKTQPYTVSEVVQVIQLRAAVERVNLSPSA 142
ATNRG++T+RGT+ + G+P+DFLDRLLII TQPYT E+ ++++R E V ++ A
Sbjct 325 ATNRGMTTIRGTNQISAHGIPIDFLDRLLIITTQPYTQDEIRNILEIRCQEEDVEMNEEA 384
Query 143 LEALGEIGGQISVRFALQLLEPCRLAAEARGSEVVEPNDVADVDSLYLDAKASPVRLAEQ 202
+ L IG S+R+A+ L+ LA R +VVE D+ V L+LD K S L E
Sbjct 385 KQLLTLIGCNTSLRYAIHLINAAALACLKRKGKVVEIQDIERVYRLFLDTKRSMQYLVEH 444
Query 203 PHRFPF 208
+ F
Sbjct 445 ESEYLF 450
> SPBC83.08
Length=465
Score = 179 bits (453), Expect = 5e-45, Method: Compositional matrix adjust.
Identities = 85/184 (46%), Positives = 124/184 (67%), Gaps = 0/184 (0%)
Query 23 EITERLREEINKTVNKYIEQGVAQLIPGVLFIDEVHMLDIECFSFLNRVLESPMSPIIVF 82
EI +RE+IN V+++ E+G A+++PGVLF+DEVHMLDIECFSF NR LE ++PI++
Sbjct 261 EIKPEVREQINTKVSEWREEGKAEIVPGVLFVDEVHMLDIECFSFFNRALEDDLAPIVIM 320
Query 83 ATNRGVSTVRGTDSVEPPGMPVDFLDRLLIIKTQPYTVSEVVQVIQLRAAVERVNLSPSA 142
A+NRG++ +RGT+ P G+PVD LDR+LII T PY+ EV +++++R E V++ PSA
Sbjct 321 ASNRGITRIRGTNYRSPHGIPVDLLDRMLIISTLPYSHEEVKEILKIRCQEEDVDMEPSA 380
Query 143 LEALGEIGGQISVRFALQLLEPCRLAAEARGSEVVEPNDVADVDSLYLDAKASPVRLAEQ 202
L+ L IG + S+R+AL L+ A R S +E +D+ V L+LD K S L E
Sbjct 381 LDYLSTIGQETSLRYALLLISSSNQVALKRKSATIEESDIRRVYELFLDQKRSVEYLEEY 440
Query 203 PHRF 206
+
Sbjct 441 GKNY 444
> Hs5730023
Length=463
Score = 174 bits (442), Expect = 9e-44, Method: Compositional matrix adjust.
Identities = 85/186 (45%), Positives = 119/186 (63%), Gaps = 0/186 (0%)
Query 23 EITERLREEINKTVNKYIEQGVAQLIPGVLFIDEVHMLDIECFSFLNRVLESPMSPIIVF 82
EI +RE+IN V ++ E+G A++IPGVLFIDEVHMLDIE FSFLNR LES M+P+++
Sbjct 267 EIKSEVREQINAKVAEWREEGKAEIIPGVLFIDEVHMLDIESFSFLNRALESDMAPVLIM 326
Query 83 ATNRGVSTVRGTDSVEPPGMPVDFLDRLLIIKTQPYTVSEVVQVIQLRAAVERVNLSPSA 142
ATNRG++ +RGT P G+P+D LDRLLI+ T PY+ + Q++++R E V +S A
Sbjct 327 ATNRGITRIRGTSYQSPHGIPIDLLDRLLIVSTTPYSEKDTKQILRIRCEEEDVEMSEDA 386
Query 143 LEALGEIGGQISVRFALQLLEPCRLAAEARGSEVVEPNDVADVDSLYLDAKASPVRLAEQ 202
L IG + S+R+A+QL+ L R V+ +D+ V SL+LD S + E
Sbjct 387 YTVLTRIGLETSLRYAIQLITAASLVCRKRKGTEVQVDDIKRVYSLFLDESRSTQYMKEY 446
Query 203 PHRFPF 208
F F
Sbjct 447 QDAFLF 452
> 7293815
Length=481
Score = 174 bits (441), Expect = 1e-43, Method: Compositional matrix adjust.
Identities = 85/186 (45%), Positives = 120/186 (64%), Gaps = 0/186 (0%)
Query 23 EITERLREEINKTVNKYIEQGVAQLIPGVLFIDEVHMLDIECFSFLNRVLESPMSPIIVF 82
EI + +R++IN V ++ E+G A++ PGVLFIDEVHMLDIECFSFLNR LES M+P++V
Sbjct 263 EIKQEVRDQINNKVLEWREEGKAEINPGVLFIDEVHMLDIECFSFLNRALESDMAPVVVM 322
Query 83 ATNRGVSTVRGTDSVEPPGMPVDFLDRLLIIKTQPYTVSEVVQVIQLRAAVERVNLSPSA 142
ATNRG++ +RGT+ P G+P+D LDR++II+T PY+ EV +++++R E + P A
Sbjct 323 ATNRGITRIRGTNYRSPHGIPIDLLDRMIIIRTVPYSEKEVKEILKIRCEEEDCIMHPDA 382
Query 143 LEALGEIGGQISVRFALQLLEPCRLAAEARGSEVVEPNDVADVDSLYLDAKASPVRLAEQ 202
L L I S+R+A+QL+ L R + V DV V SL+LD S L E
Sbjct 383 LTILTRIATDTSLRYAIQLITTANLVCRRRKATEVNTEDVKKVYSLFLDENRSSKILKEY 442
Query 203 PHRFPF 208
+ F
Sbjct 443 QDDYMF 448
> CE08426
Length=458
Score = 174 bits (441), Expect = 1e-43, Method: Compositional matrix adjust.
Identities = 92/207 (44%), Positives = 125/207 (60%), Gaps = 19/207 (9%)
Query 1 RHQQVGTDVLSMMGAYMKPRKTEITERLREEINKTVNKYIEQGVAQLIPGVLFIDEVHML 60
R Q DV +++ M P+KTE+T+RLR EINK VN+YIE GVA+L+PGVLFIDEVHML
Sbjct 267 RPQGRQGDVSNIVSQLMTPKKTEVTDRLRSEINKVVNEYIESGVAELMPGVLFIDEVHML 326
Query 61 DIECFSFLNRVLESPMSPIIVFATNRGVSTVRGTDSVEPPGMPVDFLDRLLIIKTQPYTV 120
D+ECF++L R LESPM+P++VFATNRG +TVRG P G+P + LDRL+II T Y
Sbjct 327 DVECFTYLYRALESPMAPVVVFATNRGTTTVRGLGDKAPHGIPPEMLDRLMIIPTMKYNE 386
Query 121 SEVVQVIQLRAAVERVNLSPSALEALGEIGGQISVRFALQLLEPCRLAAEARGSEVVEPN 180
++ +++ R E V A + L RL A+ G EV+E
Sbjct 387 EDIRKILVHRTEAENVQFEEKAFDLL------------------TRLCAQTCGREVIEVE 428
Query 181 DVADVDSLYLDAKASPVRLAEQPHRFP 207
DV L++D + ++ AE+ R P
Sbjct 429 DVDRCTKLFMD-RGESLKKAEEEMRQP 454
> YPL235w
Length=471
Score = 162 bits (409), Expect = 5e-40, Method: Compositional matrix adjust.
Identities = 80/184 (43%), Positives = 116/184 (63%), Gaps = 0/184 (0%)
Query 23 EITERLREEINKTVNKYIEQGVAQLIPGVLFIDEVHMLDIECFSFLNRVLESPMSPIIVF 82
EI +R++IN V ++ E+G A+++PGVLFIDEVHMLDIECFSF+NR LE +PI++
Sbjct 264 EIRSEVRDQINTKVAEWKEEGKAEIVPGVLFIDEVHMLDIECFSFINRALEDEFAPIVMM 323
Query 83 ATNRGVSTVRGTDSVEPPGMPVDFLDRLLIIKTQPYTVSEVVQVIQLRAAVERVNLSPSA 142
ATNRGVS RGT+ P G+P+D LDR +II T+ Y E+ ++ +RA E V LS A
Sbjct 324 ATNRGVSKTRGTNYKSPHGLPLDLLDRSIIITTKSYNEQEIKTILSIRAQEEEVELSSDA 383
Query 143 LEALGEIGGQISVRFALQLLEPCRLAAEARGSEVVEPNDVADVDSLYLDAKASPVRLAEQ 202
L+ L + G + S+R++ L+ + A R + VE DV L+LD+ S + E
Sbjct 384 LDLLTKTGVETSLRYSSNLISVAQQIAMKRKNNTVEVEDVKRAYLLFLDSARSVKYVQEN 443
Query 203 PHRF 206
++
Sbjct 444 ESQY 447
> CE17254
Length=448
Score = 156 bits (394), Expect = 3e-38, Method: Compositional matrix adjust.
Identities = 77/184 (41%), Positives = 112/184 (60%), Gaps = 0/184 (0%)
Query 23 EITERLREEINKTVNKYIEQGVAQLIPGVLFIDEVHMLDIECFSFLNRVLESPMSPIIVF 82
EI +R++INK V ++ E+G A+ +PGVLFIDE HMLDIECFSFLNR +E +SP+I+
Sbjct 263 EIKAEVRDQINKKVLEWREEGKAKFVPGVLFIDEAHMLDIECFSFLNRAIEGELSPLIIM 322
Query 83 ATNRGVSTVRGTDSVEPPGMPVDFLDRLLIIKTQPYTVSEVVQVIQLRAAVERVNLSPSA 142
ATNR + VRGTD G+P DFLDR+LII PYT + +++ +R E V L P+A
Sbjct 323 ATNRLIEKVRGTDVESAHGIPSDFLDRMLIINAIPYTKEDTAKILSIRCDEEGVKLQPTA 382
Query 143 LEALGEIGGQISVRFALQLLEPCRLAAEARGSEVVEPNDVADVDSLYLDAKASPVRLAEQ 202
L+ L ++ S+R+ + L+ + +E+V + + L+ D K S L E+
Sbjct 383 LDLLVKLQEATSLRYCIHLIAASEVIRIRSKAEIVTTDHIGSAYRLFFDTKRSEKILTEE 442
Query 203 PHRF 206
F
Sbjct 443 SAGF 446
> ECU09g1390
Length=426
Score = 143 bits (360), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 68/158 (43%), Positives = 108/158 (68%), Gaps = 1/158 (0%)
Query 6 GTDVLSMMGAYMKPRKTEITERLREEINKTVNKYIEQGVAQLIPGVLFIDEVHMLDIECF 65
G D+LS++ + PRKTEITERLR ++N+ VN Y+E G A+++PGVLFIDEVHMLD+ECF
Sbjct 244 GQDMLSLVFRILSPRKTEITERLRGDVNRMVNGYLENGNAEIVPGVLFIDEVHMLDVECF 303
Query 66 SFLNRVLESPMSPIIVFATNRGVSTVRGTDSVEPP-GMPVDFLDRLLIIKTQPYTVSEVV 124
+FL++V+ESP+SP I+FA+N+G++ ++G+D + P G+ D LDR++II +
Sbjct 304 TFLHKVIESPLSPTIIFASNKGMAPIKGSDGLLGPFGITKDLLDRIVIISVKRNPDEANR 363
Query 125 QVIQLRAAVERVNLSPSALEALGEIGGQISVRFALQLL 162
++I+ R E + + A + S+R+ + L+
Sbjct 364 EIIRRRMKEEGLEMDDDAFGFFVGLSTSRSLRYCISLI 401
> ECU11g1270
Length=418
Score = 121 bits (303), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 65/169 (38%), Positives = 98/169 (57%), Gaps = 1/169 (0%)
Query 23 EITERLREEINKTVNKYIEQGVAQLIPGVLFIDEVHMLDIECFSFLNRVLESPMSPIIVF 82
EI R+E+NK V +I +G A+++ GVLFIDEVHMLDIE F+FLN+ +E P+I+
Sbjct 241 EIRAETRDEVNKKVWGWINEGKAEIVRGVLFIDEVHMLDIESFAFLNKAVEEDFCPVILV 300
Query 83 ATNRGVSTVRGTDSVEPPGMPVDFLDRLLIIKTQPYTVSEVVQVIQLRAAVERVNLSPSA 142
+TN+G VRGTD P G+P DF+DR LII + + ++ +++ R E + + A
Sbjct 301 STNKGECIVRGTDEPSPYGIPRDFIDRALIISMEKHCRRDLEAILRHRILEEDILIDDDA 360
Query 143 LEALGEIGGQISVRFALQLLEPCRLAAEARGSEVVEPNDVADVDSLYLD 191
++ L I +R+++ LL + A R V DV L+LD
Sbjct 361 VDRLVSISEASGLRYSMNLLTISSMRASRRNGRVA-LGDVERAFELFLD 408
> 7299675
Length=2059
Score = 35.4 bits (80), Expect = 0.086, Method: Compositional matrix adjust.
Identities = 26/100 (26%), Positives = 53/100 (53%), Gaps = 7/100 (7%)
Query 30 EEINKTVNKYIEQGVAQLIPGVLFIDEVHMLDIECFSFLNRVLESPMSPIIVFATNRG-- 87
E+ N VNK +EQG + I G++ +DEVH++ + + +LE ++ I+ + G
Sbjct 333 EKANSIVNKLMEQGKLETI-GMVVVDEVHLISDKGRGY---ILELLLAKILYMSRRNGLQ 388
Query 88 VSTVRGTDSVEPPGMPVDFLD-RLLIIKTQPYTVSEVVQV 126
+ + + ++E + +LD L I +P + E+++V
Sbjct 389 IQVITMSATLENVQLLQSWLDAELYITNYRPVALKEMIKV 428
> Hs18105000
Length=689
Score = 32.0 bits (71), Expect = 0.85, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 32/64 (50%), Gaps = 0/64 (0%)
Query 33 NKTVNKYIEQGVAQLIPGVLFIDEVHMLDIECFSFLNRVLESPMSPIIVFATNRGVSTVR 92
+KT G Q + +++ D++ FS +N E P+S VF+T++ VST +
Sbjct 588 DKTFPPSFPSGTIQQKQQTQVKNPLNLADLDMFSSVNCSSEKPLSFSAVFSTSKSVSTPQ 647
Query 93 GTDS 96
T S
Sbjct 648 STGS 651
> Hs18105002
Length=401
Score = 31.6 bits (70), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 15/42 (35%), Positives = 27/42 (64%), Gaps = 0/42 (0%)
Query 55 DEVHMLDIECFSFLNRVLESPMSPIIVFATNRGVSTVRGTDS 96
+ +++ D++ FS +N E P+S VF+T++ VST + T S
Sbjct 322 NPLNLADLDMFSSVNCSSEKPLSFSAVFSTSKSVSTPQSTGS 363
> Hs8051621
Length=400
Score = 31.2 bits (69), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 20/68 (29%), Positives = 32/68 (47%), Gaps = 4/68 (5%)
Query 28 LREEINKTVNKYIEQGVAQLIPGVLFIDEV-HMLDIECFSFLNRVLESPMSPIIVFATNR 86
LR K+++K+IE L+P F ++ H +DI C R P+ V +
Sbjct 4 LRNTPAKSLDKFIED---YLLPDTCFRMQIDHAIDIICGFLKERCFRGSSYPVCVSKVVK 60
Query 87 GVSTVRGT 94
G S+ +GT
Sbjct 61 GGSSGKGT 68
> ECU08g1900
Length=2832
Score = 30.8 bits (68), Expect = 1.8, Method: Composition-based stats.
Identities = 23/85 (27%), Positives = 37/85 (43%), Gaps = 7/85 (8%)
Query 50 GVLFIDEVHMLDIECFSFLNRVLES------PMSPIIVFATNRGVSTVRGTDSVEPPGM- 102
G + +DE+++ LN VL+ P + V R +T+ DS M
Sbjct 1302 GWIILDEINLCTQSVIEGLNSVLDYRRKLFIPEVTVDVHEDTRIFATLNPCDSRNGRKML 1361
Query 103 PVDFLDRLLIIKTQPYTVSEVVQVI 127
P FLDR + IK YT ++ ++
Sbjct 1362 PKSFLDRFVRIKMDGYTTDDITHIL 1386
> Hs8051623
Length=364
Score = 30.8 bits (68), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 20/68 (29%), Positives = 32/68 (47%), Gaps = 4/68 (5%)
Query 28 LREEINKTVNKYIEQGVAQLIPGVLFIDEV-HMLDIECFSFLNRVLESPMSPIIVFATNR 86
LR K+++K+IE L+P F ++ H +DI C R P+ V +
Sbjct 4 LRNTPAKSLDKFIED---YLLPDTCFRMQINHAIDIICGFLKERCFRGSSYPVCVSKVVK 60
Query 87 GVSTVRGT 94
G S+ +GT
Sbjct 61 GGSSGKGT 68
> Hs7382454
Length=520
Score = 30.8 bits (68), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 27/88 (30%), Positives = 42/88 (47%), Gaps = 4/88 (4%)
Query 92 RGTDSVEPPGMPVDFLDRLLIIKTQPYTVSEV-VQVIQLRA-AVERVNLSPSALEALGEI 149
R S+ P D+ + LI ++Q Y+ +E+ +Q+ Q +E + + P A GE
Sbjct 150 RSACSISVTYFPFDWQNCSLIFQSQTYSTNEIDLQLSQEDGQTIEWIFIDPEAFTENGEW 209
Query 150 GGQISVRFALQLLEPCRLAAEARGSEVV 177
I R A LL+P A EA +VV
Sbjct 210 A--IQHRPAKMLLDPAAPAQEAGHQKVV 235
> At3g24170
Length=499
Score = 30.0 bits (66), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 20/71 (28%), Positives = 38/71 (53%), Gaps = 5/71 (7%)
Query 73 ESPMSPIIVFATNRGVSTVRGTDSVEPPGMPVDFLDRLLIIKTQPYTVSEVVQVIQLRAA 132
E ++ +++FAT R +T R ++E G+ LD+ +K Y+ + + + + A
Sbjct 285 EEFVADVVLFATGRSPNTKR--LNLEAVGVE---LDQAGAVKVDEYSRTNIPSIWAVGDA 339
Query 133 VERVNLSPSAL 143
R+NL+P AL
Sbjct 340 TNRINLTPVAL 350
> CE01305
Length=681
Score = 29.3 bits (64), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 22/79 (27%), Positives = 36/79 (45%), Gaps = 3/79 (3%)
Query 20 RKTEITERLREEINKTVNKYIEQGVAQL-IPGVLFIDEVHMLDIECFSFLNRVLESPMSP 78
RK + R+R K++ +E V+ +E +D++ SF NRV E P +
Sbjct 303 RKLQCCGRMRCAQGKSM--VVEDNCCYCNTAAVVLANEARNIDLQFMSFRNRVYEVPFAV 360
Query 79 IIVFATNRGVSTVRGTDSV 97
I V T+RG+ S+
Sbjct 361 IADHDKKSIVITIRGSCSL 379
> At4g32700
Length=1548
Score = 28.9 bits (63), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 29/48 (60%), Gaps = 3/48 (6%)
Query 14 GAYMKPRKTEITERLREEINKTVNKYIEQG-VAQLIPGVLFIDEVHML 60
G P+ T + E+ N +N+ +E+G +++L G++ IDE+HM+
Sbjct 48 GGGTLPKDTSVAVCTIEKANSLINRLLEEGRLSEL--GIIVIDELHMV 93
Lambda K H
0.320 0.137 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3718707824
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40