bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0345_orf3
Length=235
Score E
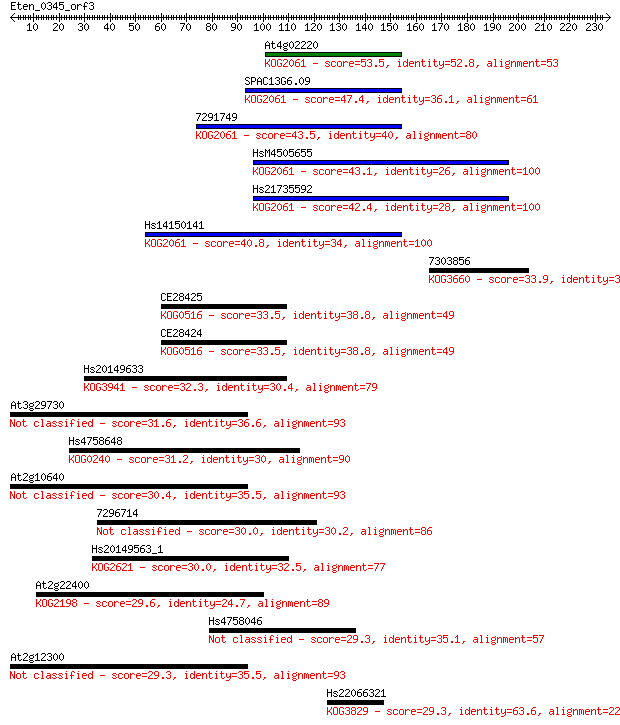
Sequences producing significant alignments: (Bits) Value
At4g02220 53.5 4e-07
SPAC13G6.09 47.4 3e-05
7291749 43.5 3e-04
HsM4505655 43.1 5e-04
Hs21735592 42.4 8e-04
Hs14150141 40.8 0.002
7303856 33.9 0.27
CE28425 33.5 0.38
CE28424 33.5 0.39
Hs20149633 32.3 0.80
At3g29730 31.6 1.6
Hs4758648 31.2 2.1
At2g10640 30.4 3.2
7296714 30.0 4.5
Hs20149563_1 30.0 4.7
At2g22400 29.6 5.1
Hs4758046 29.3 7.1
At2g12300 29.3 7.6
Hs22066321 29.3 8.2
> At4g02220
Length=446
Score = 53.5 bits (127), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 28/56 (50%), Positives = 39/56 (69%), Gaps = 5/56 (8%)
Query 101 SFLQRVRRHPKRGQVLRYAKAGG--PLWPFSQQQLGPAEVPRC-SCGGRRQFEFQV 153
+F QRV + P+ QVLRY+++ G PLWP + ++ +E+P C SCGG R FEFQV
Sbjct 342 NFQQRVDKAPE--QVLRYSRSSGAKPLWPIASGRVSKSELPSCKSCGGPRCFEFQV 395
> SPAC13G6.09
Length=274
Score = 47.4 bits (111), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 22/63 (34%), Positives = 34/63 (53%), Gaps = 4/63 (6%)
Query 93 AGGDRQLESFLQRVRRHPKRGQVLRYAKAGGPLWP--FSQQQLGPAEVPRCSCGGRRQFE 150
A GD F +R+ R P Q++RY A +P + + P+ +P C+CG +RQ E
Sbjct 156 AKGDVSFLKFQKRLSRAP--DQIMRYYHATSNEFPGLWCNNECIPSSIPNCACGAKRQLE 213
Query 151 FQV 153
FQ+
Sbjct 214 FQI 216
> 7291749
Length=347
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 32/107 (29%), Positives = 49/107 (45%), Gaps = 31/107 (28%)
Query 74 EEELDESELAAFEELHAE---------------------TAGGDRQLESFLQRVRRHPKR 112
E++ DE+ LA F+EL + A D+ F ++ P
Sbjct 197 EDKDDEARLAEFQELESSGKTGDLSNVSEAEMDKYFGNSAAADDKTFRQFKKQTAAEP-- 254
Query 113 GQVLRYAKAGGPLWPFS-----QQQLGPAEVPRC-SCGGRRQFEFQV 153
Q++RY + G PLW + + QL ++P C +CGG RQFEFQ+
Sbjct 255 DQIVRYKRGGQPLWITNTVKTVEDQLN--KLPNCIACGGERQFEFQI 299
> HsM4505655
Length=344
Score = 43.1 bits (100), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 26/108 (24%), Positives = 47/108 (43%), Gaps = 10/108 (9%)
Query 96 DRQLESFLQRVRRHPKRGQVLRYAKAGGPLWPFSQQQLGPAEVPRCSCGGRRQFEFQVGD 155
D+ + F ++ P+ Q+LRY + P+W + ++P C CG +R EFQV
Sbjct 238 DKIFQKFKTQIALEPE--QILRYGRGIAPIWISGENIPQEKDIPDCPCGAKRILEFQVMP 295
Query 156 R-------RQFGETV-WGDSLGFRSCLETVLGDSFAFTRCWSRRLGET 195
+ + G+++ WG F LG + W + + +T
Sbjct 296 QLLNYLKADRLGKSIDWGILAAFTCAESCSLGTGYTEEFVWKQDVTDT 343
> Hs21735592
Length=344
Score = 42.4 bits (98), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 28/109 (25%), Positives = 51/109 (46%), Gaps = 12/109 (11%)
Query 96 DRQLESFLQRVRRHPKRGQVLRYAKAGGPLWPFSQQQLGPAEVPRCSCGGRRQFEFQVGD 155
D+ + F ++ P+ Q+LRY + P+W + ++P C CG +R EFQV
Sbjct 238 DKIFQKFKTQIALEPE--QILRYGRGIAPIWISGENIPQEKDIPDCPCGAKRILEFQVMP 295
Query 156 R-------RQFGETV-WGDSLGFRSCLETV-LGDSFAFTRCWSRRLGET 195
+ + G+++ WG L +C E+ LG + W + + +T
Sbjct 296 QLLNYLKADRLGKSIDWG-ILAVFTCAESCSLGTGYTEEFVWKQDVTDT 343
> Hs14150141
Length=358
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 34/106 (32%), Positives = 55/106 (51%), Gaps = 12/106 (11%)
Query 54 KLSHEEQLLLNYHRNVTQHPEEELDESELAAFEELHAETA--GGDRQLESFLQRVRRHPK 111
L H LL +Y + ++ L +S +E + +T GD+ F++R+
Sbjct 203 NLDHAHSLLRDYQQREGIAMDQLLSQSLPNDGDEKYEKTIIKSGDQTFYKFMKRIAAC-- 260
Query 112 RGQVLRYAKAGGPLW---PFSQQQLGPAEVPRCS-CGGRRQFEFQV 153
+ Q+LRY+ +G PL+ P S+ E+P CS CGG+R FEFQ+
Sbjct 261 QEQILRYSWSGEPLFLTCPTSEV----TELPACSQCGGQRIFEFQL 302
> 7303856
Length=651
Score = 33.9 bits (76), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 13/39 (33%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 165 GDSLGFRSCLETVLGDSFAFTRCWSRRLGETVWGHSLGF 203
G ++G SC+ TV+ D F + W+ +G + G+ LG
Sbjct 432 GSNVGMASCMSTVIKDQFGHLKNWTVVVGIAIVGYFLGL 470
> CE28425
Length=3574
Score = 33.5 bits (75), Expect = 0.38, Method: Composition-based stats.
Identities = 19/49 (38%), Positives = 25/49 (51%), Gaps = 0/49 (0%)
Query 60 QLLLNYHRNVTQHPEEELDESELAAFEELHAETAGGDRQLESFLQRVRR 108
Q+L N +V Q P + DE + EEL+ AGG QL+ Q RR
Sbjct 243 QMLQNDVEDVDQDPRFQRDEDRIQRIEELNRMAAGGSSQLDDAEQASRR 291
> CE28424
Length=3563
Score = 33.5 bits (75), Expect = 0.39, Method: Composition-based stats.
Identities = 19/49 (38%), Positives = 25/49 (51%), Gaps = 0/49 (0%)
Query 60 QLLLNYHRNVTQHPEEELDESELAAFEELHAETAGGDRQLESFLQRVRR 108
Q+L N +V Q P + DE + EEL+ AGG QL+ Q RR
Sbjct 243 QMLQNDVEDVDQDPRFQRDEDRIQRIEELNRMAAGGSSQLDDAEQASRR 291
> Hs20149633
Length=431
Score = 32.3 bits (72), Expect = 0.80, Method: Compositional matrix adjust.
Identities = 24/80 (30%), Positives = 36/80 (45%), Gaps = 6/80 (7%)
Query 30 CTDTAEKETLEALLRRLQKGDSSSKLSHE-EQLLLNYHRNVTQHPEEELDESELAAFEEL 88
C ++ + RRL +G S +H EQ L+ Q P + L FE+L
Sbjct 21 CGAALTGTSISQVPRRLPRGLHCSAAAHSSEQSLVPSPPEPRQRPTKAL-----VPFEDL 75
Query 89 HAETAGGDRQLESFLQRVRR 108
+ GG+R SFLQ V++
Sbjct 76 FGQAPGGERDKASFLQTVQK 95
> At3g29730
Length=517
Score = 31.6 bits (70), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 34/93 (36%), Positives = 48/93 (51%), Gaps = 9/93 (9%)
Query 1 PRCLTLLKFGSVFVSFKESPLTAETEELGCTDTAEKETLEALLRRLQKGDSSSKLSHEEQ 60
PR LTL ++ ++F+ E ELGC KETL + R+L GDS+ K + EE+
Sbjct 383 PRDLTLDEYAAIFLESTEKDARGNPYELGCL----KETLGSANRQL-PGDSTFK-ALEER 436
Query 61 LLLNYHRNVTQHPEEELDESELAAFEELHAETA 93
L + Q E ++E+AA E AETA
Sbjct 437 LQEAQRQIEEQAAYNEKRDAEIAARE---AETA 466
> Hs4758648
Length=963
Score = 31.2 bits (69), Expect = 2.1, Method: Composition-based stats.
Identities = 27/98 (27%), Positives = 47/98 (47%), Gaps = 9/98 (9%)
Query 24 ETEELGCTDTAEKETLEALLRRLQKGDSSSK------LSHEEQLLLNYHRNVTQHPEEEL 77
+ EEL + +++ ++A L RLQ + +SK L E+L +NY + +Q E++
Sbjct 455 DQEELLASTRRDQDNMQAELNRLQAENDASKEEVKEVLQALEELAVNYDQK-SQEVEDKT 513
Query 78 DESELAAFE--ELHAETAGGDRQLESFLQRVRRHPKRG 113
E EL + E + A A D +L+ + KR
Sbjct 514 KEYELLSDELNQKSATLASIDAELQKLKEMTNHQKKRA 551
> At2g10640
Length=400
Score = 30.4 bits (67), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 33/94 (35%), Positives = 47/94 (50%), Gaps = 11/94 (11%)
Query 1 PRCLTLLKFGSVFVSFKESPLTAETEELGCTDTAEKETLEALLRRLQKGDSSSKLSHEEQ 60
PR LTL ++ ++F+ E LGC KETL + R+L GDS+ K E+
Sbjct 266 PRDLTLDEYAAIFLESTEKDARGNPYGLGCL----KETLGSANRQL-PGDSTFKAL--EE 318
Query 61 LLLNYHRNVTQHPE-EELDESELAAFEELHAETA 93
L R + +H E ++E+AA E AETA
Sbjct 319 RLQKAQRQIEEHAAYNEKRDAEIAARE---AETA 349
> 7296714
Length=951
Score = 30.0 bits (66), Expect = 4.5, Method: Composition-based stats.
Identities = 26/86 (30%), Positives = 41/86 (47%), Gaps = 6/86 (6%)
Query 35 EKETLEALLRRLQKGDSSSKLSHEEQLLLNYHRNVTQHPEEELDESELAAFEELHAETAG 94
+KE+L LR+L++ D+ +L + Q L RN+ E+ EL L E
Sbjct 510 DKESLGYELRKLRESDTLKELQDQRQNLATVQRNLQL---AEMKSEELKKL--LETEKLS 564
Query 95 GDRQLESFLQRVRRHPKRGQVLRYAK 120
+R L++ QR R KR + + AK
Sbjct 565 HERDLQALRQRSERE-KREEAVAVAK 589
> Hs20149563_1
Length=305
Score = 30.0 bits (66), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 25/78 (32%), Positives = 39/78 (50%), Gaps = 2/78 (2%)
Query 33 TAEKETLEALLRRLQKGDSSSKLSHEEQLLLNYHRNVTQHPEEELDESELAAFEELH-AE 91
TA+ +ALL+R + KL +QLLL + +VT+ E+D ELA L +
Sbjct 169 TAQNAMTKALLKRPLREIQMEKLKISDQLLLEIN-DVTRAWGLEVDRVELAVEAVLQPPQ 227
Query 92 TAGGDRQLESFLQRVRRH 109
+ L+S LQ++ H
Sbjct 228 DSPAGPNLDSTLQQLALH 245
> At2g22400
Length=837
Score = 29.6 bits (65), Expect = 5.1, Method: Composition-based stats.
Identities = 22/91 (24%), Positives = 41/91 (45%), Gaps = 2/91 (2%)
Query 11 SVFVSFKESPLTAETEELGCTDTAEKETLEALLRRLQKG-DSSSKLSHEEQLLLNYHRNV 69
S+ F+E P T TD+ EK + + + G S + + N +N
Sbjct 507 SLLTEFQEKPNTKRNSTAKSTDSTEKSPSKESVVTVDAGVPDESAVEKVIEADSNIEKND 566
Query 70 TQHPEEELDESE-LAAFEELHAETAGGDRQL 99
+ PE+++ E E + +E ++ AGG R++
Sbjct 567 SLEPEKKITEGESITEDKEANSSNAGGKRKV 597
> Hs4758046
Length=360
Score = 29.3 bits (64), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 20/59 (33%), Positives = 26/59 (44%), Gaps = 2/59 (3%)
Query 79 ESELAAF--EELHAETAGGDRQLESFLQRVRRHPKRGQVLRYAKAGGPLWPFSQQQLGP 135
E+EL A E H + GG Q + + H +L Y + GPL P S LGP
Sbjct 197 EAELQAVLHPEFHIQFLGGKSQKDQNTFQKLLHINATALLEYPEYSGPLLPASAVPLGP 255
> At2g12300
Length=307
Score = 29.3 bits (64), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 33/93 (35%), Positives = 47/93 (50%), Gaps = 9/93 (9%)
Query 1 PRCLTLLKFGSVFVSFKESPLTAETEELGCTDTAEKETLEALLRRLQKGDSSSKLSHEEQ 60
PR LTL ++ ++F+ E LGC KETL + R+L GDS+ K + EE+
Sbjct 224 PRDLTLDEYAAIFLESTEKDARGNPYGLGCL----KETLGSANRQL-PGDSTFK-ALEER 277
Query 61 LLLNYHRNVTQHPEEELDESELAAFEELHAETA 93
L + Q E ++E+AA E AETA
Sbjct 278 LQEAQRQIEEQAAYNEKRDAEIAARE---AETA 307
> Hs22066321
Length=541
Score = 29.3 bits (64), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 14/23 (60%), Positives = 15/23 (65%), Gaps = 1/23 (4%)
Query 125 LWPFSQQQLGPAEVPR-CSCGGR 146
LWP Q+QL P E PR C C GR
Sbjct 27 LWPQVQRQLRPRERPRGCPCTGR 49
Lambda K H
0.322 0.136 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4633705180
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40