bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0352_orf1
Length=143
Score E
Sequences producing significant alignments: (Bits) Value
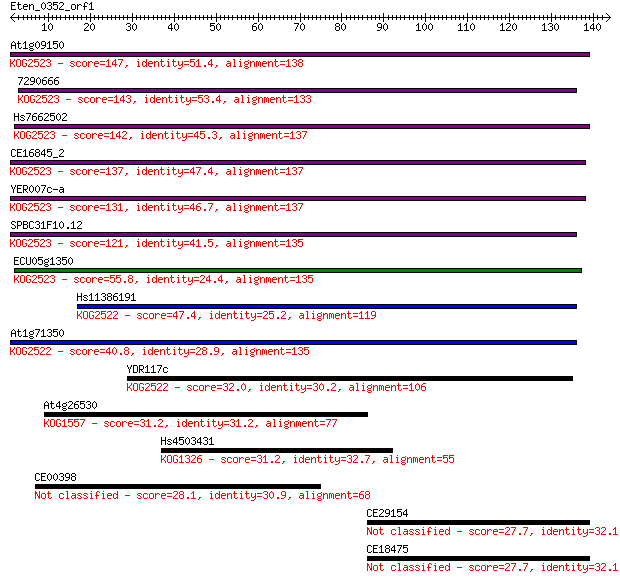
At1g09150 147 8e-36
7290666 143 1e-34
Hs7662502 142 2e-34
CE16845_2 137 5e-33
YER007c-a 131 4e-31
SPBC31F10.12 121 4e-28
ECU05g1350 55.8 2e-08
Hs11386191 47.4 8e-06
At1g71350 40.8 9e-04
YDR117c 32.0 0.39
At4g26530 31.2 0.63
Hs4503431 31.2 0.64
CE00398 28.1 5.5
CE29154 27.7 7.6
CE18475 27.7 7.6
> At1g09150
Length=181
Score = 147 bits (370), Expect = 8e-36, Method: Compositional matrix adjust.
Identities = 71/139 (51%), Positives = 97/139 (69%), Gaps = 1/139 (0%)
Query 1 EQIWPKKASPVLLKCQNRISLVVLDGKALFFQHRDRQWVPTLRLLHEYPSMMPKMQVDRG 60
E + PKK +++KC N ++LVV++ LFF RD ++PTLRLLH+YP++M + QVDRG
Sbjct 40 EDLLPKKIPLIVVKCPNHLTLVVVNNVPLFFCIRDGPYMPTLRLLHQYPNIMKRFQVDRG 99
Query 61 AVKFVLRGSNVMCQGLTSPGGRME-EVPANTVVQIAVEGKELSCAIGITTMSTDEIRREN 119
A+KFVL G+N+MC GLTSPGG ++ EV A V I EGK+ + AIG T MS +I+ N
Sbjct 100 AIKFVLSGANIMCPGLTSPGGVLDQEVEAERPVAIYAEGKQHALAIGFTKMSAKDIKSIN 159
Query 120 KGPCIETLTSLNDGLWSFE 138
KG ++ + LNDGLW E
Sbjct 160 KGIGVDNMHYLNDGLWKME 178
> 7290666
Length=182
Score = 143 bits (361), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 71/134 (52%), Positives = 91/134 (67%), Gaps = 1/134 (0%)
Query 3 IWPKKASPVLLKCQNRISLVVLD-GKALFFQHRDRQWVPTLRLLHEYPSMMPKMQVDRGA 61
I PKK S + KC + I L++ G +FF+HRD W+PTLRLLH++P + QVD+GA
Sbjct 43 ILPKKDSYRIAKCHDHIELLLNGAGDQVFFRHRDGPWMPTLRLLHKFPYFVTMQQVDKGA 102
Query 62 VKFVLRGSNVMCQGLTSPGGRMEEVPANTVVQIAVEGKELSCAIGITTMSTDEIRRENKG 121
++FVL G+NVMC GLTSPG M +TVV I EGKE + A+G+ T+ST EI +NKG
Sbjct 103 IRFVLSGANVMCPGLTSPGACMTPADKDTVVAIMAEGKEHALAVGLLTLSTQEILAKNKG 162
Query 122 PCIETLTSLNDGLW 135
IET LNDGLW
Sbjct 163 IGIETYHFLNDGLW 176
> Hs7662502
Length=181
Score = 142 bits (358), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 62/137 (45%), Positives = 93/137 (67%), Gaps = 0/137 (0%)
Query 2 QIWPKKASPVLLKCQNRISLVVLDGKALFFQHRDRQWVPTLRLLHEYPSMMPKMQVDRGA 61
QI PKK +++C I ++ ++G+ LFF+ R+ + PTLRLLH+YP ++P QVD+GA
Sbjct 42 QIMPKKDPVKIVRCHEHIEILTVNGELLFFRQREGPFYPTLRLLHKYPFILPHQQVDKGA 101
Query 62 VKFVLRGSNVMCQGLTSPGGRMEEVPANTVVQIAVEGKELSCAIGITTMSTDEIRRENKG 121
+KFVL G+N+MC GLTSPG ++ +T+V I EGK+ + +G+ MS ++I + NKG
Sbjct 102 IKFVLSGANIMCPGLTSPGAKLYPAAVDTIVAIMAEGKQHALCVGVMKMSAEDIEKVNKG 161
Query 122 PCIETLTSLNDGLWSFE 138
IE + LNDGLW +
Sbjct 162 IGIENIHYLNDGLWHMK 178
> CE16845_2
Length=180
Score = 137 bits (346), Expect = 5e-33, Method: Compositional matrix adjust.
Identities = 65/139 (46%), Positives = 96/139 (69%), Gaps = 2/139 (1%)
Query 1 EQIWPKKASPVLLKCQNRISLVVLD-GKALFFQHRDRQWVPTLRLLHEYPSMMPKMQVDR 59
E+I PKK + ++KC++ I L+ G F + R+ ++PTLR LH+YP ++P QVD+
Sbjct 36 EEILPKKENFKVIKCKDHIELLADHLGVVRFVKTRNTDYIPTLRTLHKYPFILPHQQVDK 95
Query 60 GAVKFVLRGSNVMCQGLTSPGGRMEE-VPANTVVQIAVEGKELSCAIGITTMSTDEIRRE 118
GA+KF+L GS++MC GLTSPG ++ VP ++VV + EGK+ + AIG+ +MS++EI+
Sbjct 96 GAIKFILNGSSIMCPGLTSPGAKLTPLVPKDSVVAVMAEGKQHALAIGLMSMSSEEIQSV 155
Query 119 NKGPCIETLTSLNDGLWSF 137
NKG IE L LNDGLW
Sbjct 156 NKGNGIENLHYLNDGLWHL 174
> YER007c-a
Length=181
Score = 131 bits (330), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 64/140 (45%), Positives = 92/140 (65%), Gaps = 4/140 (2%)
Query 1 EQIWPKKASPVLLKCQNRISLVVLDGKALFFQHRDRQWVPTLRLLHEYPSMMPKMQVDRG 60
+++ PKK+ L+KC+++I L +DG+ LFFQ D + +P+L+L+H++P P +QVDRG
Sbjct 40 DELIPKKSQIELIKCEDKIQLYSVDGEVLFFQKFD-ELIPSLKLVHKFPEAYPTVQVDRG 98
Query 61 AVKFVLRGSNVMCQGLTSPGGRMEEVPA---NTVVQIAVEGKELSCAIGITTMSTDEIRR 117
A+KFVL G+N+MC GLTS G + P T+V I E KE + AIG M T+EI+
Sbjct 99 AIKFVLSGANIMCPGLTSAGADLPPAPGYEKGTIVVINAENKENALAIGELMMGTEEIKS 158
Query 118 ENKGPCIETLTSLNDGLWSF 137
NKG IE + L D LW+F
Sbjct 159 VNKGHSIELIHHLGDPLWNF 178
> SPBC31F10.12
Length=184
Score = 121 bits (304), Expect = 4e-28, Method: Compositional matrix adjust.
Identities = 56/136 (41%), Positives = 88/136 (64%), Gaps = 1/136 (0%)
Query 1 EQIWPKKASPVLLKCQNRISLVVLDGKALFFQHRDRQWVPTLRLLHEYPSMMPKMQVDRG 60
+++ PKK+ +KC++R+ L L+G+ + FQH D +P+LRL+H+ P +++VDRG
Sbjct 44 DELIPKKSQLTQIKCEDRLFLYTLNGEIILFQHFDGPIIPSLRLVHKCPDAFTQVRVDRG 103
Query 61 AVKFVLRGSNVMCQGLTSPGGRM-EEVPANTVVQIAVEGKELSCAIGITTMSTDEIRREN 119
A+KF+L G+N+M GL S GG + +++ + V + EGKE AIG+T MS E++ N
Sbjct 104 AIKFLLSGANIMIPGLVSKGGNLPDDIEKDQYVIVTAEGKEAPAAIGLTKMSAKEMKETN 163
Query 120 KGPCIETLTSLNDGLW 135
KG IE + L D LW
Sbjct 164 KGIGIENVHYLGDNLW 179
> ECU05g1350
Length=203
Score = 55.8 bits (133), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 33/136 (24%), Positives = 73/136 (53%), Gaps = 3/136 (2%)
Query 2 QIWPKKASPVLLKCQNRISLVVLDGKALFFQHRDRQWVPTLRLLHEYPSMMPKMQVDRGA 61
++ K + ++ +C+N+ SL+ L G A+ F +R++ PT+R L + ++ +D GA
Sbjct 66 EVLEKNETYMVHRCKNKASLISLGGSAILF-CLNRKYFPTIRYLEGHGGGFYEVFLDEGA 124
Query 62 VKFVLRGSNVMCQGLTSPGGRMEE-VPANTVVQIAVEGKELSCAIGITTMSTDEIRRENK 120
++ + RG++VM G+ +++ A + + + GK + A+G + ++ ++
Sbjct 125 LEPLCRGADVMIPGVLKYKDLIKKGFEAGDTIVVRIIGKSI-VAVGEAIVGLKDMEASSR 183
Query 121 GPCIETLTSLNDGLWS 136
G IE + DGL++
Sbjct 184 GVGIEVYHRVGDGLYT 199
> Hs11386191
Length=584
Score = 47.4 bits (111), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 30/120 (25%), Positives = 56/120 (46%), Gaps = 2/120 (1%)
Query 17 NRISLVVLDGKALFFQHRDRQWVPTLRLLHEYPSMMPKMQVDRGAVKFVLRGSNVMCQGL 76
+ +++ V G + F+ ++ PT+ L YP ++P ++ ++ G+++M GL
Sbjct 59 DAVTVYVSGGNPILFE-LEKNLYPTVYTLWSYPDLLPTFTTWPLVLEKLVGGADLMLPGL 117
Query 77 TSPGGRMEEVPANTVVQIAVEGKELSCAIGITTMSTDEIRREN-KGPCIETLTSLNDGLW 135
P + +V + I++ G AIG+ MST E+ KG L + D LW
Sbjct 118 VMPPAGLPQVQKGDLCAISLVGNRAPVAIGVAAMSTAEMLTSGLKGRGFSVLHTYQDHLW 177
> At1g71350
Length=566
Score = 40.8 bits (94), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 39/139 (28%), Positives = 58/139 (41%), Gaps = 26/139 (18%)
Query 1 EQIWPKKASPVLLKCQNRISLVVLDGK-ALFFQ--HRDRQWVPTLRLLHEYPSMMPKMQV 57
+ I P K L K QNR+ + ++G +FF R + PT+ L E P M+P
Sbjct 39 DAILPPKVEITLSKFQNRVIVYSIEGGCPMFFDIDGRGTEIFPTVFALWEAPEMLPS--- 95
Query 58 DRGAVKFVLRGSNVMCQGLTSPGGRMEEVPANTVVQIAVEGKELSCAIGITTMSTDEIRR 117
F+L+G G S A + + V G A+G TTMS++E R+
Sbjct 96 ------FMLKG------GYPS-------FSAGEIWAVKVPGNLAPIAVGCTTMSSEEARK 136
Query 118 EN-KGPCIETLTSLNDGLW 135
+G + D LW
Sbjct 137 AGLRGKALRITHYYRDFLW 155
> YDR117c
Length=565
Score = 32.0 bits (71), Expect = 0.39, Method: Composition-based stats.
Identities = 32/112 (28%), Positives = 53/112 (47%), Gaps = 10/112 (8%)
Query 29 LFFQHRDRQWVPTLRLLHEYPSMMPKMQVDRGAV--KFVLRGSNVMCQGLTSPGGRMEEV 86
LF + Q PT+ EYP+++P + + G V + + G+N+M G P ++
Sbjct 66 LFKEKHKEQLFPTVYSCWEYPALLP-IVLTHGFVIEEHLFNGANLMISGSIPPFDPRCKI 124
Query 87 PANTVVQIAV-EGKELSCAIGITTM---STDEIRRENKGPCIETLTSLNDGL 134
T+ IA + E AIGI + S D++ E G ++ + NDGL
Sbjct 125 --GTLCGIASKQAPETVLAIGIVELDLPSFDKVIGET-GVAVKIIHHFNDGL 173
> At4g26530
Length=358
Score = 31.2 bits (69), Expect = 0.63, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 37/77 (48%), Gaps = 6/77 (7%)
Query 9 SPVLLKCQNRISLVVLDGKALFFQHRDRQWVPTLRLLHEYPSMMPKMQVDRGAVKFVLRG 68
SP C +S V+L + L+ + D + P + LL E ++P ++VD+G V
Sbjct 62 SPGTFPC---LSGVILFEETLYQKTTDGK--PFVELLME-NGVIPGIKVDKGVVDLAGTN 115
Query 69 SNVMCQGLTSPGGRMEE 85
QGL S G R +E
Sbjct 116 GETTTQGLDSLGARCQE 132
> Hs4503431
Length=2080
Score = 31.2 bits (69), Expect = 0.64, Method: Compositional matrix adjust.
Identities = 18/60 (30%), Positives = 30/60 (50%), Gaps = 5/60 (8%)
Query 37 QWVPTLRLLHEYPSMMPKMQV-----DRGAVKFVLRGSNVMCQGLTSPGGRMEEVPANTV 91
QW + L +PSM KM++ DR ++ + + +++PGG +EE PA V
Sbjct 441 QWNQNITLPAMFPSMCEKMRIRIIDWDRLTHNDIVATTYLSMSKISAPGGEIEEEPAGAV 500
> CE00398
Length=164
Score = 28.1 bits (61), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 21/68 (30%), Positives = 33/68 (48%), Gaps = 3/68 (4%)
Query 7 KASPVLLKCQNRISLVVLDGKALFFQHRDRQWVPTLRLLHEYPSMMPKMQVDRGAVKFVL 66
K SPV KC+ V + +F + RQ + T++ L K+Q + GA++F L
Sbjct 16 KESPV--KCEYSDEKKVSECLQVFLYFKTRQCL-TMKKLQPMLDYATKLQAETGAMQFPL 72
Query 67 RGSNVMCQ 74
+G V Q
Sbjct 73 QGGQVFNQ 80
> CE29154
Length=564
Score = 27.7 bits (60), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 17/54 (31%), Positives = 31/54 (57%), Gaps = 4/54 (7%)
Query 86 VPANTVVQIAVEGKELSCAIGITTMSTDEIRRENKGPCIETLTSLNDGL-WSFE 138
VP +T +I V+ L+ +GIT+ TD+I EN P + + + ++ L W+ +
Sbjct 484 VPRSTDEKIRVK---LTSPVGITSDQTDQINSENAEPNVGIILNKDNNLEWTIK 534
> CE18475
Length=531
Score = 27.7 bits (60), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 17/54 (31%), Positives = 31/54 (57%), Gaps = 4/54 (7%)
Query 86 VPANTVVQIAVEGKELSCAIGITTMSTDEIRRENKGPCIETLTSLNDGL-WSFE 138
VP +T +I V+ L+ +GIT+ TD+I EN P + + + ++ L W+ +
Sbjct 451 VPRSTDEKIRVK---LTSPVGITSDQTDQINSENAEPNVGIILNKDNNLEWTIK 501
Lambda K H
0.321 0.136 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1639544670
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40