bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0357_orf1
Length=192
Score E
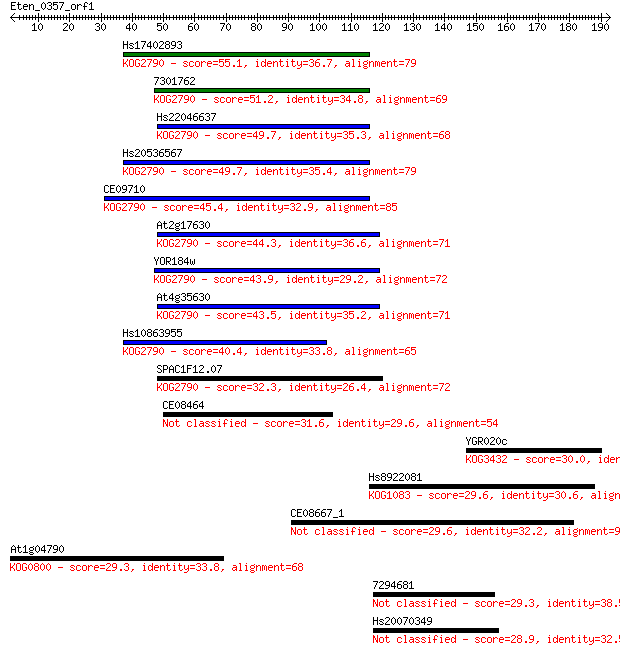
Sequences producing significant alignments: (Bits) Value
Hs17402893 55.1 8e-08
7301762 51.2 1e-06
Hs22046637 49.7 3e-06
Hs20536567 49.7 4e-06
CE09710 45.4 7e-05
At2g17630 44.3 2e-04
YOR184w 43.9 2e-04
At4g35630 43.5 3e-04
Hs10863955 40.4 0.002
SPAC1F12.07 32.3 0.63
CE08464 31.6 1.1
YGR020c 30.0 2.7
Hs8922081 29.6 3.8
CE08667_1 29.6 3.9
At1g04790 29.3 4.4
7294681 29.3 5.2
Hs20070349 28.9 7.1
> Hs17402893
Length=370
Score = 55.1 bits (131), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 29/80 (36%), Positives = 45/80 (56%), Gaps = 1/80 (1%)
Query 37 SELPERTLAADSCL-NTPPALNIYILGKVLQHLAAAGDLNYWDGVCCSKSAAVYRHCDCS 95
S L + A +S L NTPP +IY++G VL+ + G + + KS +Y D S
Sbjct 226 SVLEYKVQAGNSSLYNTPPCFSIYVMGLVLEWIKNNGGAAAMEKLSSIKSQTIYEIIDNS 285
Query 96 SGFFLAPVKPQFRSRVAVRF 115
GF++ PV+PQ RS++ + F
Sbjct 286 QGFYVCPVEPQNRSKMNIPF 305
> 7301762
Length=364
Score = 51.2 bits (121), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 24/69 (34%), Positives = 37/69 (53%), Gaps = 0/69 (0%)
Query 47 DSCLNTPPALNIYILGKVLQHLAAAGDLNYWDGVCCSKSAAVYRHCDCSSGFFLAPVKPQ 106
+S LNTPP IY++G V + + G + + +KS +Y + S+GF+ PV
Sbjct 231 NSLLNTPPTFGIYVMGLVFKWIKRNGGVAGMAKLAAAKSKLIYDTINQSNGFYYCPVDVN 290
Query 107 FRSRVAVRF 115
RSR+ V F
Sbjct 291 VRSRMNVPF 299
> Hs22046637
Length=481
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 24/68 (35%), Positives = 37/68 (54%), Gaps = 0/68 (0%)
Query 48 SCLNTPPALNIYILGKVLQHLAAAGDLNYWDGVCCSKSAAVYRHCDCSSGFFLAPVKPQF 107
S NTPP +IYI+G +L+ G + + KS +Y D S GF++ PV+ Q
Sbjct 249 SSYNTPPCFSIYIMGLLLEWTKNNGGAAAMEKLSSIKSQMIYEITDNSQGFYVCPVETQN 308
Query 108 RSRVAVRF 115
RS++ + F
Sbjct 309 RSKMNIPF 316
> Hs20536567
Length=189
Score = 49.7 bits (117), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 28/80 (35%), Positives = 42/80 (52%), Gaps = 1/80 (1%)
Query 37 SELPERTLAADSCL-NTPPALNIYILGKVLQHLAAAGDLNYWDGVCCSKSAAVYRHCDCS 95
S L + A +S L N PP +IY+ G VL+ + G + + KS +Y D S
Sbjct 10 SVLEYKVQAGNSSLYNMPPCFSIYVRGLVLESIKNNGGVAAMEKRSSIKSQMIYEIIDNS 69
Query 96 SGFFLAPVKPQFRSRVAVRF 115
GF++ PV PQ RS++ + F
Sbjct 70 QGFYICPVGPQNRSKMNIPF 89
> CE09710
Length=370
Score = 45.4 bits (106), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 28/85 (32%), Positives = 41/85 (48%), Gaps = 4/85 (4%)
Query 31 PSAXSVSELPERTLAADSCLNTPPALNIYILGKVLQHLAAAGDLNYWDGVCCSKSAAVYR 90
PS S E+ +A +S NTPP IY VL+ + + G L + KS +Y
Sbjct 224 PSVFSYKEM----IANNSLYNTPPTGGIYTTNLVLKWIKSKGGLQAIYELNLQKSGMIYD 279
Query 91 HCDCSSGFFLAPVKPQFRSRVAVRF 115
D S+GF+ V ++RS + V F
Sbjct 280 IIDNSNGFYHCAVDKRYRSIMNVCF 304
> At2g17630
Length=422
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 26/71 (36%), Positives = 33/71 (46%), Gaps = 0/71 (0%)
Query 48 SCLNTPPALNIYILGKVLQHLAAAGDLNYWDGVCCSKSAAVYRHCDCSSGFFLAPVKPQF 107
S NTPP IY+ G V L G L + K+ +Y D S GFF PV+
Sbjct 295 SLYNTPPCFGIYMCGLVFDDLLEQGGLKEVEKKNQRKAELLYNAIDESRGFFRCPVEKSV 354
Query 108 RSRVAVRFLLQ 118
RS + V F L+
Sbjct 355 RSLMNVPFTLE 365
> YOR184w
Length=395
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 21/72 (29%), Positives = 36/72 (50%), Gaps = 0/72 (0%)
Query 47 DSCLNTPPALNIYILGKVLQHLAAAGDLNYWDGVCCSKSAAVYRHCDCSSGFFLAPVKPQ 106
+S NT P ++++ V QH+ G + K+ +Y D +S F+ PV P+
Sbjct 267 NSAYNTIPIFTLHVMDLVFQHILKKGGVEAQQAENEEKAKILYEALDANSDFYNVPVDPK 326
Query 107 FRSRVAVRFLLQ 118
RS++ V F L+
Sbjct 327 CRSKMNVVFTLK 338
> At4g35630
Length=430
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 35/71 (49%), Gaps = 0/71 (0%)
Query 48 SCLNTPPALNIYILGKVLQHLAAAGDLNYWDGVCCSKSAAVYRHCDCSSGFFLAPVKPQF 107
S NTPP IY+ G V + L G L + K+ +Y + S+GFF PV+
Sbjct 303 SLYNTPPCFGIYMCGLVFEDLLEQGGLKEVEKKNQRKADLLYNAIEESNGFFRCPVEKSV 362
Query 108 RSRVAVRFLLQ 118
RS + V F L+
Sbjct 363 RSLMNVPFTLE 373
> Hs10863955
Length=324
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 35/66 (53%), Gaps = 1/66 (1%)
Query 37 SELPERTLAADSCL-NTPPALNIYILGKVLQHLAAAGDLNYWDGVCCSKSAAVYRHCDCS 95
S L + A +S L NTPP +IY++G VL+ + G + + KS +Y D S
Sbjct 226 SVLEYKVQAGNSSLYNTPPCFSIYVMGLVLEWIKNNGGAAAMEKLSSIKSQTIYEIIDNS 285
Query 96 SGFFLA 101
GF+++
Sbjct 286 QGFYVS 291
> SPAC1F12.07
Length=389
Score = 32.3 bits (72), Expect = 0.63, Method: Compositional matrix adjust.
Identities = 19/72 (26%), Positives = 34/72 (47%), Gaps = 1/72 (1%)
Query 48 SCLNTPPALNIYILGKVLQHLAAAGDLNYWDGVCCSKSAAVYRHCDCSSGFFLAPVKPQF 107
S NT P ++ + L+++ G L + KS +Y D +++ V+P
Sbjct 262 SLYNTLPVATLHAINLGLEYMLEHGGLVALEASSIEKSKLLYDTLD-KHDLYISVVEPAA 320
Query 108 RSRVAVRFLLQP 119
RSR+ V F ++P
Sbjct 321 RSRMNVTFRIEP 332
> CE08464
Length=244
Score = 31.6 bits (70), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 16/54 (29%), Positives = 27/54 (50%), Gaps = 2/54 (3%)
Query 50 LNTPPALNIYILGKVLQHLAAAGDLNYWDGVCCSKSAAVYRHCDCSSGFFLAPV 103
L+TPP +N + + +Q A G++ + DG + S + C G +APV
Sbjct 104 LSTPPTVNFTFVPQKVQ--IADGEVCFTDGTSIAISGTIVYKCSIGGGSTVAPV 155
> YGR020c
Length=118
Score = 30.0 bits (66), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 23/43 (53%), Gaps = 0/43 (0%)
Query 147 CTPEKEKENSRVLQEAKLKDEELTKVFLKRAEGRNMICLVHTN 189
TPE +++N V QE K EE+T F E R+ I ++ N
Sbjct 28 ITPETQEKNFFVYQEGKTTKEEITDKFNHFTEERDDIAILLIN 70
> Hs8922081
Length=2969
Score = 29.6 bits (65), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 34/75 (45%), Gaps = 13/75 (17%)
Query 116 LLQPHAAAAAAAAADCSKREPSESSQPDAAVCTPEKEKENSRVLQEAKLKDEELTKVFLK 175
L +P A A ++ K+ ESSQ + CTPE+ + N R E L ++ L
Sbjct 2881 LEEPEREGATANVSEGEKKT-EESSQEPQSTCTPEERRHNQR---------ERLNQILLN 2930
Query 176 RAE---GRNMICLVH 187
E G+N I + +
Sbjct 2931 LLEKIPGKNAIDVTY 2945
> CE08667_1
Length=415
Score = 29.6 bits (65), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 29/102 (28%), Positives = 41/102 (40%), Gaps = 13/102 (12%)
Query 91 HCDCSSGFFLAPV------KPQFR-SRVAVRFLLQPHAAAAAAAAADCSKREPSESSQPD 143
H + S F PV K Q + +R AV L+Q H A+ D P D
Sbjct 276 HTEVQSSFGATPVEIVAQNKEQCQEARNAVMSLMQSHQDKPASNPPDSGFSTPGSPFTSD 335
Query 144 AAVCTPEKEKENSRVLQEAKLKDEE-----LTKVFLKRAEGR 180
++ TPEK + NSR +D+ LT F+ + R
Sbjct 336 SSSTTPEK-RGNSRQYHRGSFRDQPKVMLALTPQFIDKEHCR 376
> At1g04790
Length=612
Score = 29.3 bits (64), Expect = 4.4, Method: Composition-based stats.
Identities = 23/78 (29%), Positives = 33/78 (42%), Gaps = 16/78 (20%)
Query 1 IVNXDLFFXEMGFLEXSARCXLLLQX---RQGGPSAXSVSELPERTLAADS-------CL 50
I + +L + F E L L R GG SA ++ LPE T+ D+ CL
Sbjct 512 ITSSNLLHMDRDFTEDDYELLLALDENNHRHGGASANRINNLPESTVQTDNFQETCVICL 571
Query 51 NTPPALNIYILGKVLQHL 68
TP +G ++HL
Sbjct 572 ETPK------IGDTIRHL 583
> 7294681
Length=1121
Score = 29.3 bits (64), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 15/39 (38%), Positives = 17/39 (43%), Gaps = 0/39 (0%)
Query 117 LQPHAAAAAAAAADCSKREPSESSQPDAAVCTPEKEKEN 155
L PH AA AA +C + PD VC E K N
Sbjct 837 LGPHLAAVTLAAYECQTLDFPTPPYPDIKVCPQEPYKRN 875
> Hs20070349
Length=406
Score = 28.9 bits (63), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 13/40 (32%), Positives = 19/40 (47%), Gaps = 0/40 (0%)
Query 117 LQPHAAAAAAAAADCSKREPSESSQPDAAVCTPEKEKENS 156
L PH A+ AA +C+ E PD +C E+ E +
Sbjct 138 LGPHIASVTLAAYECNSVNFPEPPYPDQIICPDEEGTEGT 177
Lambda K H
0.319 0.132 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3238438140
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40