bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0358_orf2
Length=186
Score E
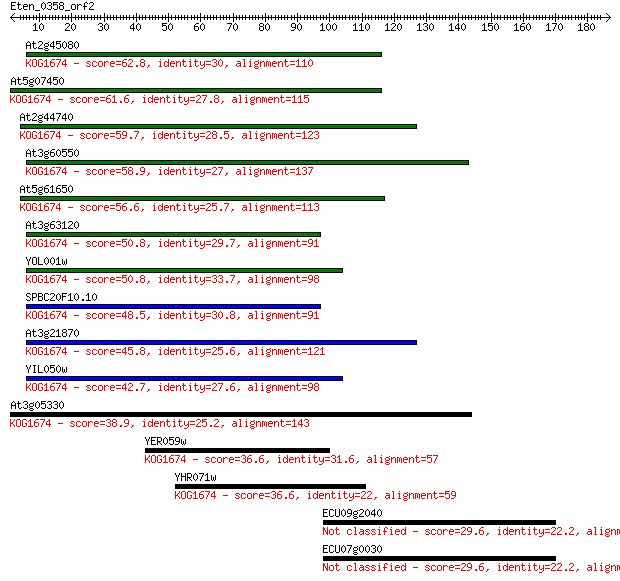
Sequences producing significant alignments: (Bits) Value
At2g45080 62.8 4e-10
At5g07450 61.6 8e-10
At2g44740 59.7 4e-09
At3g60550 58.9 5e-09
At5g61650 56.6 3e-08
At3g63120 50.8 1e-06
YOL001w 50.8 2e-06
SPBC20F10.10 48.5 7e-06
At3g21870 45.8 5e-05
YIL050w 42.7 4e-04
At3g05330 38.9 0.006
YER059w 36.6 0.028
YHR071w 36.6 0.029
ECU09g2040 29.6 3.6
ECU07g0030 29.6 3.6
> At2g45080
Length=222
Score = 62.8 bits (151), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 33/110 (30%), Positives = 53/110 (48%), Gaps = 0/110 (0%)
Query 6 PPWTLGQYVTRLQHLASATEHEMLMALLLLTHLQQRQQQLRISQKNAHRLLLAALVLVSK 65
P T+ Y+ R+ A ++A + + Q Q RIS N HRLL+ +++ SK
Sbjct 67 PDMTIQSYLERIFRYTKAGPSVYVVAYVYIDRFCQNNQGFRISLTNVHRLLITTIMIASK 126
Query 66 VVRDDHTPLAMWAVVGGVPPRELAALELALLELVGHRVSFTFPEFAAAYC 115
V D + + +A VGG+ +L LEL L L+G ++ F + C
Sbjct 127 YVEDMNYKNSYFAKVGGLETEDLNNLELEFLFLMGFKLHVNVSVFESYCC 176
> At5g07450
Length=216
Score = 61.6 bits (148), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 32/115 (27%), Positives = 60/115 (52%), Gaps = 1/115 (0%)
Query 1 NSSTEPPWTLGQYVTRLQHLASATEHEMLMALLLLTHLQQRQQQLRISQKNAHRLLLAAL 60
N+ T+P ++ Y+ R+ A ++ ++A + L Q+Q L I N HRL++ ++
Sbjct 55 NAVTKPSISIRSYMERIFKYADCSDSCYIVAYIYLDRFIQKQPLLPIDSSNVHRLIITSV 114
Query 61 VLVSKVVRDDHTPLAMWAVVGGVPPRELAALELALLELVGHRVSFTFPEFAAAYC 115
++ +K + D A +A VGG+ E+ LEL L +G +++ T + YC
Sbjct 115 LVSAKFMDDLCYNNAFYAKVGGITTEEMNLLELDFLFGIGFQLNVTISTY-NDYC 168
> At2g44740
Length=202
Score = 59.7 bits (143), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 35/123 (28%), Positives = 64/123 (52%), Gaps = 6/123 (4%)
Query 4 TEPPWTLGQYVTRLQHLASATEHEMLMALLLLTHLQQRQQQLRISQKNAHRLLLAALVLV 63
+ P T+ Y+ R+ A+ + ++A + L RQ L I+ N HRLL+ ++++
Sbjct 48 SRPTITIQSYLERIFKYANCSPSCFVVAYVYLDRFTHRQPSLPINSFNVHRLLITSVMVA 107
Query 64 SKVVRDDHTPLAMWAVVGGVPPRELAALELALLELVGHRVSFTFPEFAAAYCLAAAVSHL 123
+K + D + A +A VGG+ +E+ LEL L +G ++ T F A + S+L
Sbjct 108 AKFLDDLYYNNAYYAKVGGISTKEMNFLELDFLFGLGFELNVTPNTFNAYF------SYL 161
Query 124 QQQ 126
Q++
Sbjct 162 QKE 164
> At3g60550
Length=230
Score = 58.9 bits (141), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 37/142 (26%), Positives = 65/142 (45%), Gaps = 5/142 (3%)
Query 6 PPWTLGQYVTRLQHLASATEHEMLMALLLLTHLQQRQQQLRISQKNAHRLLLAALVLVSK 65
P T+ Y+ R+ A ++A + + Q RIS N HRLL+ +++ SK
Sbjct 73 PDMTIQSYLGRIFRYTKAGPSVYVVAYVYIDRFCQTNPGFRISLTNVHRLLITTIMIASK 132
Query 66 VVRDDHTPLAMWAVVGGVPPRELAALELALLELVGHRVSFTFPEFAAAYC-----LAAAV 120
V D + + +A VGG+ +L LEL L L+G ++ F + C ++
Sbjct 133 YVEDLNYRNSYFAKVGGLETEDLNKLELEFLFLMGFKLHVNVSVFESYCCHLEREVSFGG 192
Query 121 SHLQQQQRRPSAQQKQRQQLPQ 142
+ ++ R + + K RQ + Q
Sbjct 193 GYQIEKALRCAEEIKSRQMIIQ 214
> At5g61650
Length=219
Score = 56.6 bits (135), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 29/113 (25%), Positives = 56/113 (49%), Gaps = 0/113 (0%)
Query 4 TEPPWTLGQYVTRLQHLASATEHEMLMALLLLTHLQQRQQQLRISQKNAHRLLLAALVLV 63
T+P ++ Y+ R+ A+ + ++A + L ++Q L I+ N HRL++ ++++
Sbjct 56 TKPSISIRSYLERIFEYANCSYSCYIVAYIYLDRFVKKQPFLPINSFNVHRLIITSVLVS 115
Query 64 SKVVRDDHTPLAMWAVVGGVPPRELAALELALLELVGHRVSFTFPEFAAAYCL 116
+K + D +A VGG+ E+ LEL L +G ++ T F C
Sbjct 116 AKFMDDLSYNNEYYAKVGGISREEMNMLELDFLFGIGFELNVTVSTFNNYCCF 168
> At3g63120
Length=221
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 27/91 (29%), Positives = 48/91 (52%), Gaps = 1/91 (1%)
Query 6 PPWTLGQYVTRLQHLASATEHEMLMALLLLTHLQQRQQQLRISQKNAHRLLLAALVLVSK 65
P ++ Y+ R+ + + ++A + + H + + L + N HRL++ ++L +K
Sbjct 71 PEISIAHYLDRIFKYSCCSPSCFVIAHIYIDHFLHKTRAL-LKPLNVHRLIITTVMLAAK 129
Query 66 VVRDDHTPLAMWAVVGGVPPRELAALELALL 96
V D + A +A VGGV REL LE+ LL
Sbjct 130 VFDDRYFNNAYYARVGGVTTRELNRLEMELL 160
> YOL001w
Length=293
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 33/99 (33%), Positives = 49/99 (49%), Gaps = 2/99 (2%)
Query 6 PPWTLGQYVTRLQHLASATEHEMLM-ALLLLTHLQQRQQQLRISQKNAHRLLLAALVLVS 64
P ++ Y RL +S EH +LM +L + LQ ++ AHR LL A + +
Sbjct 73 PNISIFNYFIRLTKFSSL-EHCVLMTSLYYIDLLQTVYPDFTLNSLTAHRFLLTATTVAT 131
Query 65 KVVRDDHTPLAMWAVVGGVPPRELAALELALLELVGHRV 103
K + D + A +A VGGV EL LE L+ V +R+
Sbjct 132 KGLCDSFSTNAHYAKVGGVRCHELNILENDFLKRVNYRI 170
> SPBC20F10.10
Length=243
Score = 48.5 bits (114), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 28/92 (30%), Positives = 41/92 (44%), Gaps = 1/92 (1%)
Query 6 PPWTLGQYVTRLQHLASATEHEMLMALLLLTHL-QQRQQQLRISQKNAHRLLLAALVLVS 64
P ++ Y+TR+ AT L L+ L + + I+ N HR L+A S
Sbjct 78 PSISIQAYLTRILKYCPATNDVFLSVLIYLDRIVHHFHFTVFINSFNIHRFLIAGFTAAS 137
Query 65 KVVRDDHTPLAMWAVVGGVPPRELAALELALL 96
K D + +A VGG+P EL LEL+
Sbjct 138 KFFSDVFYTNSRYAKVGGIPLHELNHLELSFF 169
> At3g21870
Length=210
Score = 45.8 bits (107), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 31/121 (25%), Positives = 60/121 (49%), Gaps = 6/121 (4%)
Query 6 PPWTLGQYVTRLQHLASATEHEMLMALLLLTHLQQRQQQLRISQKNAHRLLLAALVLVSK 65
P ++ +Y+ R+ + ++ + + L + + N HRLL+ +++ +K
Sbjct 65 PSISIAKYLERIYKYTKCSPACFVVGYVYIDRLAHKHPGSLVVSLNVHRLLVTCVMIAAK 124
Query 66 VVRDDHTPLAMWAVVGGVPPRELAALELALLELVGHRVSFTFPEFAAAYCLAAAVSHLQQ 125
++ D H +A VGGV +L +EL LL L+ RV+ +F F +YC HL++
Sbjct 125 ILDDVHYNNEFYARVGGVSNADLNKMELELLFLLDFRVTVSFRVF-ESYCF-----HLEK 178
Query 126 Q 126
+
Sbjct 179 E 179
> YIL050w
Length=285
Score = 42.7 bits (99), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 27/109 (24%), Positives = 46/109 (42%), Gaps = 11/109 (10%)
Query 6 PPWTLGQYVTRLQHLASATEHEMLMALLLLTHLQQR-----------QQQLRISQKNAHR 54
P + QY+ R+Q T L L+ + + +Q + N HR
Sbjct 156 PEIAVVQYLERIQKYCPTTNDIFLSLLVYFDRISKNYGHSSERNGCAKQLFVMDSGNIHR 215
Query 55 LLLAALVLVSKVVRDDHTPLAMWAVVGGVPPRELAALELALLELVGHRV 103
LL+ + + +K + D + +A VGG+ +EL LEL L L ++
Sbjct 216 LLITGVTICTKFLSDFFYSNSRYAKVGGISLQELNHLELQFLILCDFKL 264
> At3g05330
Length=588
Score = 38.9 bits (89), Expect = 0.006, Method: Composition-based stats.
Identities = 36/147 (24%), Positives = 64/147 (43%), Gaps = 11/147 (7%)
Query 1 NSSTEPPWTLGQYVTRLQHLASATEHEMLMALLLLTHLQQRQQQL----RISQKNAHRLL 56
+ S P ++ +Y R+ A + + A + QR + R++ N HRLL
Sbjct 442 HGSKAPSLSIYRYTERIHRYAQCSPVCFVAAFAYILRYLQRPEATSTARRLTSLNVHRLL 501
Query 57 LAALVLVSKVVRDDHTPLAMWAVVGGVPPRELAALELALLELVGHRVSFTFPEFAAAYCL 116
+ +L++ +K + A +A +GGV E+ LE L V R+ T F +CL
Sbjct 502 ITSLLVAAKFLERQCYNNAYYAKIGGVSTEEMNRLERTFLVDVDFRLYITTETF-EKHCL 560
Query 117 AAAVSHLQQQQRRPSAQQKQRQQLPQL 143
+ Q++ P +K R L ++
Sbjct 561 ------MLQKETVPCDSRKLRTVLGEI 581
> YER059w
Length=420
Score = 36.6 bits (83), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 18/57 (31%), Positives = 30/57 (52%), Gaps = 0/57 (0%)
Query 43 QQLRISQKNAHRLLLAALVLVSKVVRDDHTPLAMWAVVGGVPPRELAALELALLELV 99
Q + N HRL++A + + +K + D + ++ VGG+ +EL LEL L L
Sbjct 328 QMFVMDSHNIHRLIIAGITVSTKFLSDFFYSNSRYSRVGGISLQELNHLELQFLVLC 384
> YHR071w
Length=229
Score = 36.6 bits (83), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 13/59 (22%), Positives = 31/59 (52%), Gaps = 0/59 (0%)
Query 52 AHRLLLAALVLVSKVVRDDHTPLAMWAVVGGVPPRELAALELALLELVGHRVSFTFPEF 110
+ R+ L L+L K + D+ + W ++ G+ ++L+ +E L + + ++ + EF
Sbjct 127 SRRIFLCCLILSHKFLNDNTYSMKNWQIISGLHAKDLSLMERWCLGKLNYELAIPYDEF 185
> ECU09g2040
Length=236
Score = 29.6 bits (65), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 16/72 (22%), Positives = 34/72 (47%), Gaps = 0/72 (0%)
Query 98 LVGHRVSFTFPEFAAAYCLAAAVSHLQQQQRRPSAQQKQRQQLPQLLQLPQLQQLPQQLL 157
VGH + P+ + +C + H ++ + + +++ P L+Q L+ LP +L
Sbjct 107 CVGHPLRIPSPKHSPRHCTLHDIRHRKKLHPLNNPKGANKRKTPPLIQPTVLEPLPHPVL 166
Query 158 QLRDVRSSSADP 169
++ SS+ P
Sbjct 167 SPHNISSSTDHP 178
> ECU07g0030
Length=236
Score = 29.6 bits (65), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 16/72 (22%), Positives = 34/72 (47%), Gaps = 0/72 (0%)
Query 98 LVGHRVSFTFPEFAAAYCLAAAVSHLQQQQRRPSAQQKQRQQLPQLLQLPQLQQLPQQLL 157
VGH + P+ + +C + H ++ + + +++ P L+Q L+ LP +L
Sbjct 107 CVGHPLRIPSPKHSPRHCTLHDIRHRKKLHPLNNPKGANKRKTPPLIQPTVLEPLPHPVL 166
Query 158 QLRDVRSSSADP 169
++ SS+ P
Sbjct 167 SPHNISSSTDHP 178
Lambda K H
0.319 0.130 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3022542264
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40