bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0367_orf2
Length=412
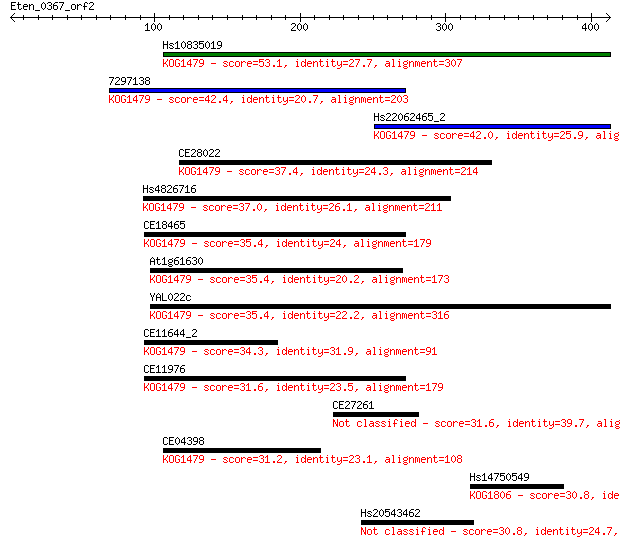
Score E
Sequences producing significant alignments: (Bits) Value
Hs10835019 53.1 1e-06
7297138 42.4 0.002
Hs22062465_2 42.0 0.003
CE28022 37.4 0.055
Hs4826716 37.0 0.084
CE18465 35.4 0.23
At1g61630 35.4 0.24
YAL022c 35.4 0.25
CE11644_2 34.3 0.53
CE11976 31.6 3.1
CE27261 31.6 3.5
CE04398 31.2 4.4
Hs14750549 30.8 5.0
Hs20543462 30.8 5.1
> Hs10835019
Length=456
Score = 53.1 bits (126), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 85/334 (25%), Positives = 143/334 (42%), Gaps = 49/334 (14%)
Query 106 NGALQSSGFARAAILPRNYVGTTSIGQAVAGLVAFVVTAILMNGVFDMDKEID-VKWFCS 164
+ LQ S F + +P Y GQ +AG+ F A+L++ +D E + +F +
Sbjct 141 SAVLQGSLFGQLGTMPSTYSTLFLSGQGLAGI--FAALAMLLSMASGVDAETSALGYFIT 198
Query 165 ICCGISV-LLCVLSMLYMHF--FLNAKVCVQSVNNALEGK-----SDAKTVPDDTPTSVA 216
GI + ++C LS+ ++ F + A Q+ LE K SD +P +P VA
Sbjct 199 PYVGILMSIVCYLSLPHLKFARYYLANKSSQAQAQELETKAELLQSDENGIPS-SPQKVA 257
Query 217 -----DLE-EPKVRQSSGHASSQDESLLPPRPWL-TMVKGSWWELLSLFLVFFITFSLFP 269
DLE EP+ S DE P +P + T+ + W L L LVF +T S+FP
Sbjct 258 LTLDLDLEKEPE--------SEPDEPQKPGKPSVFTVFQKIWLTALCLVLVFTVTLSVFP 309
Query 270 KVGPVSFNFEGKSPSKMVLLFG------MEFVGDFLGRSCLTLPNLHPACSFLFLSRNGT 323
+ + SP K F + + D+LGRS + FL+ +
Sbjct 310 AI--TAMVTSSTSPGKWSQFFNPICCFLLFNIMDWLGRSLTSY--------FLWPDEDSR 359
Query 324 IIASFLRLIF-YVPFLM---AMKMENVPFI-NNFVWLMIIQLLLAFTLGWVGTLTLIHCS 378
++ + L F +VP M + +P + + + LL A + G++ +LT+
Sbjct 360 LLPLLVCLRFLFVPLFMLCHVPQRSRLPILFPQDAYFITFMLLFAVSNGYLVSLTMCLAP 419
Query 379 LSVTRVSEKARMGSVSTIVLAVAIGIGLYIALAF 412
V E+ G++ T LA+ + G ++ F
Sbjct 420 RQVLP-HEREVAGALMTFFLALGLSCGASLSFLF 452
> 7297138
Length=458
Score = 42.4 bits (98), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 42/208 (20%), Positives = 82/208 (39%), Gaps = 53/208 (25%)
Query 69 LFLSLACMAALPPAFFYGALNCRITFAHIILGLLGACNGALQSSGFARAAILPRNYVGTT 128
+ L++ + P FF+ + C + LL CNG Q++ + A LP Y G
Sbjct 151 IILAMLDSSQWPGVFFWTTMVCIV--------LLNVCNGIYQNTIYGIVASLPIKYTGAV 202
Query 129 SIGQAVAGLVAFVVTAILMNGVFDMDKEIDVKWFCS-----ICCGISVLLCVLSMLYMHF 183
+G ++G A++ +F + + +F + + C + L+ + H+
Sbjct 203 VLGSNISGCFT-TAMALICGEIFSSKRTSAIYYFVTAILVLLLCFDTYFALPLNKFFRHY 261
Query 184 FLNAKVCVQSVNNALEGKSDAKTVPDDTPTSVADLEEPKVRQSSGHASSQDESLLPPRPW 243
++++ + E KSD+K A L P+
Sbjct 262 --------ETISRSSEKKSDSK----------AQLN---------------------VPY 282
Query 244 LTMVKGSWWELLSLFLVFFITFSLFPKV 271
+ K + +L ++FL FF+T S+FP +
Sbjct 283 WQIFKKAAPQLFNIFLTFFVTLSVFPAI 310
> Hs22062465_2
Length=474
Score = 42.0 bits (97), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 42/166 (25%), Positives = 82/166 (49%), Gaps = 20/166 (12%)
Query 251 WWELLSLFLVFFITFSLFPKV-GPVSFNFEGK-SPSKMVLLFGMEFVGDFLGRSCLTLPN 308
W ++LS+ + +FIT LFP + + G+ P ++ +F + DF+G+ LP
Sbjct 298 WADMLSIAVTYFITLCLFPGLESEIRHCILGEWLPILIMAVFNLS---DFVGKILAALP- 353
Query 309 LHPACSFLFLSRNGT--IIASFLRLIFYVPFLMAMKMENVPFINNFVWLMIIQLLLAFTL 366
+ GT + S LR++F F++ + +P + + W I LL+ +
Sbjct 354 ---------VDWRGTHLLACSCLRVVFIPLFILCVYPSGMPALRHPAWPCIFSLLMGISN 404
Query 367 GWVGTLTLIHCSLSVTRVSEKARMGSVSTIVLAVAIGIGLYIALAF 412
G+ G++ +I L+ +VS K R + +T+ ++ G+ L A+A+
Sbjct 405 GYFGSVPMI---LAAGKVSPKQRELAGNTMTVSYMSGLTLGSAVAY 447
> CE28022
Length=384
Score = 37.4 bits (85), Expect = 0.055, Method: Compositional matrix adjust.
Identities = 52/219 (23%), Positives = 92/219 (42%), Gaps = 26/219 (11%)
Query 117 AAILPRNYVGTTSIGQAVAGLVAFVVTAILMNGVFDMDKEIDVKWFCSICCGISVLLCVL 176
+A+ P Y +GQ+ AG++A + +IL V DV + G S+++C +
Sbjct 156 SALFPSQYTQAVMVGQSFAGVLA-ALMSILCQAVTS-----DVILNGQMYFGFSLIMCFI 209
Query 177 SMLYMHFFLNAKVCVQSVNNALEGKSDAKTVPDDTPTSVADLEEPKVRQSSGHASSQDES 236
S L +++L ++ EG + ++ S+ S ++ +E
Sbjct 210 S-LATYYYLTTLTPPMITDDGSEG-----LIENEEEVSIEAQANHFPPIDSDNSGQTEEH 263
Query 237 LLPPRPWLT-MVKGSWWELLSLFLVFFITFSLFPK----VGPVSFNFEGKSPSKMVLLFG 291
LP T +++ S +L ++ +V +T + +P V S N S V F
Sbjct 264 QLPKWTMYTDIIRKSAIDLTTISVVLIVTLAAYPGLTSLVHSTSRNHTWNSYFSAVASFL 323
Query 292 MEFVGDFLGRSCLTLPNLHPACSFLFLSRNGTIIASFLR 330
+ VGD +GRS + + L LS +I SFLR
Sbjct 324 LYNVGDLIGRS---------SANSLRLSPKYLLIISFLR 353
> Hs4826716
Length=456
Score = 37.0 bits (84), Expect = 0.084, Method: Compositional matrix adjust.
Identities = 55/221 (24%), Positives = 90/221 (40%), Gaps = 26/221 (11%)
Query 92 ITFAHIILGLLGACNGALQSSGFARAAILPRNYVGTTSIGQAVAGLVAFVVTAILMNGVF 151
IT I+L + + LQ S F A +LP +Y GQ +AG A V +
Sbjct 142 ITMIKIVL--INSFGAILQGSLFGLAGLLPASYTAPIMSGQGLAGFFASVAMICAIASGS 199
Query 152 DMDKEIDVKWFCSICCGISV-LLCVLSMLYMHFFLNAKVCVQSVNNALEGKSDAKTVPDD 210
++ E +F + C I + ++C L + + F+ + LEG + +T D
Sbjct 200 ELS-ESAFGYFITACAVIILTIICYLGLPRLEFYRYYQ------QLKLEGPGEQETKLD- 251
Query 211 TPTSVADLEEPKV-RQSSGHASSQDESLLPPRPWLTMVKGSWWELLSLFLVFFITFSLFP 269
++ EEP+ ++ SG + S + ++K S+ +F IT +FP
Sbjct 252 ---LISKGEEPRAGKEESGVSVSNSQPTNESHSIKAILKNISVLAFSVCFIFTITIGMFP 308
Query 270 KVG-PVSFNFEGKS-------PSKMVLLFGMEFVGDFLGRS 302
V V + G S P L F + D+LGRS
Sbjct 309 AVTVEVKSSIAGSSTWERYFIPVSCFLTFN---IFDWLGRS 346
> CE18465
Length=461
Score = 35.4 bits (80), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 43/183 (23%), Positives = 74/183 (40%), Gaps = 40/183 (21%)
Query 93 TFAHIILGLLGACNGALQSSGFARAAILPRNYVGTTSIGQAVAG----LVAFVVTAILMN 148
T I + +L NG Q+S F A+ LP Y IG + G L++ A+ N
Sbjct 148 TLTIITIIVLNGANGVYQNSIFGLASELPFKYTNAVIIGNNLCGTFVTLLSMSTKAVTRN 207
Query 149 GVFDMDKEIDVKWFCSICCGISVLLCVLSMLYMHFFLNAKVCVQSVNNALEGKSDAKTVP 208
+D+ + SI I+++ C +S H L + Q + E + +
Sbjct 208 ---ILDRSFA---YFSIAL-ITLVFCFISF---HI-LKKQRFYQYYSTRAERQRNKNDEA 256
Query 209 DDTPTSVADLEEPKVRQSSGHASSQDESLLPPRPWLTMVKGSWWELLSLFLVFFITFSLF 268
D+ VA+ ++ K ++ +L+++FLVFF+T S+F
Sbjct 257 VDSEGKVAN-------------------------YIATFKEAFPQLINVFLVFFVTLSIF 291
Query 269 PKV 271
P V
Sbjct 292 PGV 294
> At1g61630
Length=382
Score = 35.4 bits (80), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 35/173 (20%), Positives = 80/173 (46%), Gaps = 10/173 (5%)
Query 97 IILGLLGACNGALQSSGFARAAILPRNYVGTTSIGQAVAGLVAFVVTAILMNGVFDMDKE 156
+I+ L G + +Q + + + +++ G +AG + V+ ++ +FD +
Sbjct 83 LIVALFGLADAFVQGAMVGDLSFMSPDFIQAFMAGLGIAGALTSVLR-LITKAIFDNSPD 141
Query 157 IDVKWFCSICCGISVLLCVLSMLYMHFFLNAKVCVQSVNNALEGKSDAKTVPDDTPTSVA 216
++ + GI+ L+ L+ ++++ + AK+ + A GK AKTV A
Sbjct 142 -GLRKGALLFIGIATLI-ELACVFLYTLVFAKLPIVKYYRAKAGKEGAKTVS-------A 192
Query 217 DLEEPKVRQSSGHASSQDESLLPPRPWLTMVKGSWWELLSLFLVFFITFSLFP 269
DL +++ + DES + +++ + ++L L++ +T S+FP
Sbjct 193 DLAAAGLQEQAEQVHQMDESKIQKLTKKQLLRENIDLGINLSLIYVVTLSIFP 245
> YAL022c
Length=517
Score = 35.4 bits (80), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 70/334 (20%), Positives = 131/334 (39%), Gaps = 24/334 (7%)
Query 97 IILGLLGACNGALQSSG-FARAAILPRNYVGTTSIGQAVAGLVAFVV---TAILMNGVFD 152
++L ++ + A+ +G A A + Y +GQAVAG++ +V A + N
Sbjct 181 MMLVVISSMGTAMTQNGIMAIANVFGSEYSQGVMVGQAVAGVLPSLVLFALAFIENSSVS 240
Query 153 MDKEIDVKWF-CSICCGISVLLCVLSMLYMHFFLNAKVCVQSVNNALEGKSDAKTVPDDT 211
I + +F ++ I V++ +S + N V + + L G +
Sbjct 241 TTGGILLYFFTTTLVVTICVVMFSVSKISRKVNENWNVEDGHITDVLLGSLRSNEEEIRI 300
Query 212 PTSVADLEEPKVRQSSGHASSQD--ESLLPPRPWLTMVKGSWWELLSLFLVFFITFSLFP 269
+ +E+ R+++G D E L P+ + + +LS+F F +T +FP
Sbjct 301 VGRIDQMEDEDHRRTNGTRDDNDEGEELQLKVPFEVLFAKLKYLVLSIFTTFVVTL-VFP 359
Query 270 KVGPVSF--NFEGKSPSKMVLLFGMEFVGDFLGRSCLTLPNLHPACSFLFLSRNGTIIAS 327
++ + + L+F + +GD GR P F R T I S
Sbjct 360 VFASATYVTGLPLSNAQYIPLIFTLWNLGDLYGRVIADWPMFRDQK---FTPRK-TFIYS 415
Query 328 FLRLIFYVPFLM---------AMKMENVPFINNFVWLMIIQLLLAFTLGWVGTLTLIHCS 378
LR+ FLM + N I + + M++Q L T G V +++ +
Sbjct 416 LLRVAAIPLFLMFTAITSSSSGDEEHNGSVIVDLCY-MLLQFLFGVTNGHVISMSFMKVP 474
Query 379 LSVTRVSEKARMGSVSTIVLAVAIGIGLYIALAF 412
+ EK G + I ++ + +G I+ F
Sbjct 475 EQLDNDDEKEAAGGFTNIFVSTGLALGSIISYVF 508
> CE11644_2
Length=737
Score = 34.3 bits (77), Expect = 0.53, Method: Compositional matrix adjust.
Identities = 29/94 (30%), Positives = 44/94 (46%), Gaps = 9/94 (9%)
Query 93 TFAHIILG---LLGACNGALQSSGFARAAILPRNYVGTTSIGQAVAGLVAFVVTAILMNG 149
TF+ + LG +L A NG Q+S F A+ P Y IGQ G A V ++L
Sbjct 436 TFSMLTLGTIVVLNAANGLFQNSMFGLASPFPFKYTNAVIIGQNFCG-TAVTVLSMLTKA 494
Query 150 VFDMDKEIDVKWFCSICCGISVLLCVLSMLYMHF 183
D DV+ S+ G+S + V+ + ++F
Sbjct 495 ASD-----DVQMRASLFFGLSSVAVVVCFILLNF 523
> CE11976
Length=450
Score = 31.6 bits (70), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 42/183 (22%), Positives = 72/183 (39%), Gaps = 40/183 (21%)
Query 93 TFAHIILGLLGACNGALQSSGFARAAILPRNYVGTTSIGQAVAG----LVAFVVTAILMN 148
T I + +L NG Q+S F A+ LP Y IG + G L++ A+ N
Sbjct 153 TLTIITIIVLNGANGVYQNSIFGLASELPFKYTNAVIIGNNLCGTFVTLLSMSTKAVTRN 212
Query 149 GVFDMDKEIDVKWFCSICCGISVLLCVLSMLYMHFFLNAKVCVQSVNNALEGKSDAKTVP 208
+D+ + SI I+++ C +S H L + Q + E +
Sbjct 213 ---ILDRSFA---YFSIAL-ITLVFCFISF---HI-LKKQRFYQFYSTRAERQRAKNEEA 261
Query 209 DDTPTSVADLEEPKVRQSSGHASSQDESLLPPRPWLTMVKGSWWELLSLFLVFFITFSLF 268
D +A+ ++ K ++ +L+++FLVFF+T S+F
Sbjct 262 ADNEGKMAN-------------------------YIATFKEAFPQLINVFLVFFVTLSIF 296
Query 269 PKV 271
P V
Sbjct 297 PGV 299
> CE27261
Length=4155
Score = 31.6 bits (70), Expect = 3.5, Method: Composition-based stats.
Identities = 23/60 (38%), Positives = 31/60 (51%), Gaps = 6/60 (10%)
Query 223 VRQSSGHASSQDESLLPPRPWLTMVKGSWWELLSLFLVFFITFSL-FPKVGPV-SFNFEG 280
+R G SS D S+L W+T KG +W + + FL F T SL F K+ FN +G
Sbjct 429 IRTRCGPLSSLDMSIL----WITTEKGFYWNMKAEFLNFEATTSLIFTKLFSCKKFNVDG 484
> CE04398
Length=434
Score = 31.2 bits (69), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 25/108 (23%), Positives = 47/108 (43%), Gaps = 8/108 (7%)
Query 106 NGALQSSGFARAAILPRNYVGTTSIGQAVAGLVAFVVTAILMNGVFDMDKEIDVKWFCSI 165
NG ++S + A P Y+G IG + GL+ VV + + D K + + +F
Sbjct 147 NGLYENSVYGVFADFPHTYIGALLIGNNICGLLITVVKIGVTYFLNDEPKLVAIVYF--- 203
Query 166 CCGISVLLCVLSMLYMHFFLNAKVCVQSVNNALEGKSDAKTVPDDTPT 213
GIS+++ ++ + + F +E + A+T D P+
Sbjct 204 --GISLVILLVCAIALFFITKQDFYHYHHQKGMEIREKAET---DRPS 246
> Hs14750549
Length=1485
Score = 30.8 bits (68), Expect = 5.0, Method: Composition-based stats.
Identities = 22/69 (31%), Positives = 33/69 (47%), Gaps = 8/69 (11%)
Query 317 FLSRNGTIIASFLRLIFYVPFLMAMKMENVPFINNFVWLMIIQLLLAFTLGWVGTL---- 372
FLS+ +I F+ ++ VP + M+ V + F+ LMI L T W T+
Sbjct 221 FLSQ---LIQKFISVLKSVPLSEPVTMDKVHYCERFIELMIDLEALLPTRRWFNTILDDS 277
Query 373 -TLIHCSLS 380
L+HC LS
Sbjct 278 HLLVHCYLS 286
> Hs20543462
Length=132
Score = 30.8 bits (68), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 19/77 (24%), Positives = 31/77 (40%), Gaps = 6/77 (7%)
Query 242 PWLTMVKGSWWELLSLFLVFFITFSLFPKVGPVSFNFEGKSPSKMVLLFGMEFVGDFLGR 301
PW T++KGS W L+ + + L + S +SP + F +FL
Sbjct 16 PWGTLLKGSLWALVRNTVYVLVALILNARPNEKSDRLGDQSPCYEI------FFNEFLYS 69
Query 302 SCLTLPNLHPACSFLFL 318
HP+C+ F+
Sbjct 70 DNRPDSGSHPSCTLAFI 86
Lambda K H
0.329 0.142 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 10524876166
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40