bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0378_orf2
Length=174
Score E
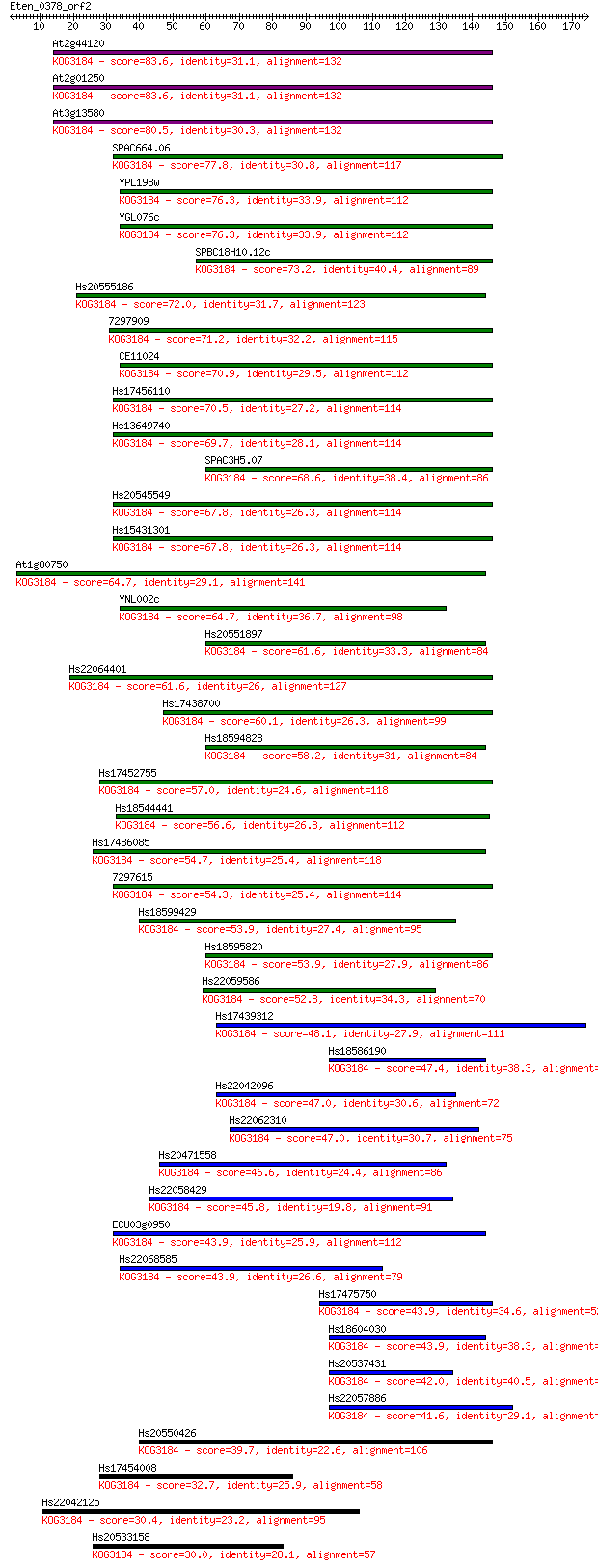
Sequences producing significant alignments: (Bits) Value
At2g44120 83.6 2e-16
At2g01250 83.6 2e-16
At3g13580 80.5 2e-15
SPAC664.06 77.8 9e-15
YPL198w 76.3 3e-14
YGL076c 76.3 3e-14
SPBC18H10.12c 73.2 2e-13
Hs20555186 72.0 6e-13
7297909 71.2 1e-12
CE11024 70.9 1e-12
Hs17456110 70.5 2e-12
Hs13649740 69.7 3e-12
SPAC3H5.07 68.6 7e-12
Hs20545549 67.8 1e-11
Hs15431301 67.8 1e-11
At1g80750 64.7 8e-11
YNL002c 64.7 9e-11
Hs20551897 61.6 7e-10
Hs22064401 61.6 8e-10
Hs17438700 60.1 2e-09
Hs18594828 58.2 8e-09
Hs17452755 57.0 2e-08
Hs18544441 56.6 2e-08
Hs17486085 54.7 1e-07
7297615 54.3 1e-07
Hs18599429 53.9 2e-07
Hs18595820 53.9 2e-07
Hs22059586 52.8 3e-07
Hs17439312 48.1 8e-06
Hs18586190 47.4 2e-05
Hs22042096 47.0 2e-05
Hs22062310 47.0 2e-05
Hs20471558 46.6 3e-05
Hs22058429 45.8 5e-05
ECU03g0950 43.9 1e-04
Hs22068585 43.9 1e-04
Hs17475750 43.9 2e-04
Hs18604030 43.9 2e-04
Hs20537431 42.0 7e-04
Hs22057886 41.6 9e-04
Hs20550426 39.7 0.003
Hs17454008 32.7 0.40
Hs22042125 30.4 2.1
Hs20533158 30.0 2.5
> At2g44120
Length=247
Score = 83.6 bits (205), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 41/135 (30%), Positives = 78/135 (57%), Gaps = 7/135 (5%)
Query 14 PECRVYCVVRNHRRGGCR-ETQEALKALGLPRPYSCTLIANDENATKILRKVSPFVFYGL 72
PE ++ ++R +T++ L+ L L + ++ + ++ +LR+V P+V YG
Sbjct 84 PEAKLLFIIRIRGINAIDPKTKKILQLLRLRQIFNGVFLKVNKATVNMLRRVEPYVTYGY 143
Query 73 PSAETIRRLFVTRA--RLTSEEGVAPVALSDNMMVEEAFGSLGLFCVEDLVHEVVTCGPH 130
P+ ++++ L R +L + +AL+DN +V++A G G+ CVEDL+HE++T GPH
Sbjct 144 PNLKSVKELIYKRGYGKLNHQR----IALTDNSIVDQALGKHGIICVEDLIHEIMTVGPH 199
Query 131 FPQVMQRVGHFQLES 145
F + + FQL++
Sbjct 200 FKEANNFLWPFQLKA 214
> At2g01250
Length=242
Score = 83.6 bits (205), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 41/135 (30%), Positives = 77/135 (57%), Gaps = 7/135 (5%)
Query 14 PECRVYCVVRNHRRGGCR-ETQEALKALGLPRPYSCTLIANDENATKILRKVSPFVFYGL 72
PE ++ ++R +T++ L+ L L + ++ + ++ +LR+V P+V YG
Sbjct 79 PEAKLLFIIRIRGINAIDPKTKKILQLLRLRQIFNGVFLKVNKATMNMLRRVEPYVTYGF 138
Query 73 PSAETIRRLFVTRA--RLTSEEGVAPVALSDNMMVEEAFGSLGLFCVEDLVHEVVTCGPH 130
P+ ++++ L R +L + +AL+DN +VE+A G G+ C EDL+HE++T GPH
Sbjct 139 PNLKSVKELIYKRGYGKLNHQR----IALTDNSIVEQALGKHGIICTEDLIHEILTVGPH 194
Query 131 FPQVMQRVGHFQLES 145
F + + FQL++
Sbjct 195 FKEANNFLWPFQLKA 209
> At3g13580
Length=244
Score = 80.5 bits (197), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 40/135 (29%), Positives = 76/135 (56%), Gaps = 7/135 (5%)
Query 14 PECRVYCVVRNHRRGGCR-ETQEALKALGLPRPYSCTLIANDENATKILRKVSPFVFYGL 72
PE ++ ++R +T++ L+ L L + ++ + ++ +LR+V P+V YG
Sbjct 81 PEAKLLFIIRIRGINAIDPKTKKILQLLRLRQIFNGVFLKVNKATINMLRRVEPYVTYGY 140
Query 73 PSAETIRRLFVTRA--RLTSEEGVAPVALSDNMMVEEAFGSLGLFCVEDLVHEVVTCGPH 130
P+ ++++ L R +L + AL+DN +V++ G G+ CVEDL+HE++T GPH
Sbjct 141 PNLKSVKELIYKRGFGKLNHQR----TALTDNSIVDQGLGKHGIICVEDLIHEIMTVGPH 196
Query 131 FPQVMQRVGHFQLES 145
F + + FQL++
Sbjct 197 FKEANNFLWPFQLKA 211
> SPAC664.06
Length=249
Score = 77.8 bits (190), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 36/119 (30%), Positives = 66/119 (55%), Gaps = 6/119 (5%)
Query 32 ETQEALKALGLPRPYSCTLIANDENATKILRKVSPFVFYGLPSAETIRRLFVTRA--RLT 89
+ ++ L+ L L R + + N++ ++LR V P+V YG+P+ ++R L R ++
Sbjct 104 KIRKVLRLLRLSRINNAVFVRNNKAVAQMLRIVEPYVMYGIPNLHSVRELIYKRGFGKIN 163
Query 90 SEEGVAPVALSDNMMVEEAFGSLGLFCVEDLVHEVVTCGPHFPQVMQRVGHFQLESLKQ 148
+ +ALSDN ++EEA G + +ED++HE+ G HF +V + + F L +K
Sbjct 164 GQR----IALSDNALIEEALGKYDVISIEDIIHEIYNVGSHFKEVTKFLWPFTLTPVKH 218
> YPL198w
Length=244
Score = 76.3 bits (186), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 38/114 (33%), Positives = 63/114 (55%), Gaps = 6/114 (5%)
Query 34 QEALKALGLPRPYSCTLIANDENATKILRKVSPFVFYGLPSAETIRRLFVTRA--RLTSE 91
++ L+ L L R S T + + ++L+ + P+V YG PS TIR+L R ++ +
Sbjct 100 RKVLQLLRLTRINSGTFVKVTKATLELLKLIEPYVAYGYPSYSTIRQLVYKRGFGKINKQ 159
Query 92 EGVAPVALSDNMMVEEAFGSLGLFCVEDLVHEVVTCGPHFPQVMQRVGHFQLES 145
V LSDN ++E G G+ ++DL+HE++T GPHF Q + F+L +
Sbjct 160 R----VPLSDNAIIEANLGKYGILSIDDLIHEIITVGPHFKQANNFLWPFKLSN 209
> YGL076c
Length=244
Score = 76.3 bits (186), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 38/114 (33%), Positives = 63/114 (55%), Gaps = 6/114 (5%)
Query 34 QEALKALGLPRPYSCTLIANDENATKILRKVSPFVFYGLPSAETIRRLFVTRA--RLTSE 91
++ L+ L L R S T + + ++L+ + P+V YG PS TIR+L R ++ +
Sbjct 100 RKVLQLLRLTRINSGTFVKVTKATLELLKLIEPYVAYGYPSYSTIRQLVYKRGFGKINKQ 159
Query 92 EGVAPVALSDNMMVEEAFGSLGLFCVEDLVHEVVTCGPHFPQVMQRVGHFQLES 145
V LSDN ++E G G+ ++DL+HE++T GPHF Q + F+L +
Sbjct 160 R----VPLSDNAIIEANLGKYGILSIDDLIHEIITVGPHFKQANNFLWPFKLSN 209
> SPBC18H10.12c
Length=251
Score = 73.2 bits (178), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 36/90 (40%), Positives = 53/90 (58%), Gaps = 3/90 (3%)
Query 57 ATK-ILRKVSPFVFYGLPSAETIRRLFVTRARLTSEEGVAPVALSDNMMVEEAFGSLGLF 115
ATK +L+ V P+V YG+P+ +T+R L R + +ALSDN ++E A G +
Sbjct 130 ATKEMLQVVEPYVTYGIPNLKTVRELLYKRG--FGKVNKQRIALSDNAIIEAALGKYSIL 187
Query 116 CVEDLVHEVVTCGPHFPQVMQRVGHFQLES 145
+EDL+HE+ T GP+F Q + FQL S
Sbjct 188 SIEDLIHEIYTVGPNFKQAANFIWPFQLSS 217
> Hs20555186
Length=255
Score = 72.0 bits (175), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 39/124 (31%), Positives = 63/124 (50%), Gaps = 3/124 (2%)
Query 21 VVRNHRRGGCRE-TQEALKALGLPRPYSCTLIANDENATKILRKVSPFVFYGLPSAETIR 79
VVR R G Q + L L + +S + K+LR V P+V +G P+ +++R
Sbjct 99 VVRIERIDGVSLLVQRTIARLRLKKIFSGVFVKVTPQNLKMLRIVEPYVTWGFPNLKSVR 158
Query 80 RLFVTRARLTSEEGVAPVALSDNMMVEEAFGSLGLFCVEDLVHEVVTCGPHFPQVMQRVG 139
L + R + + P L+DN ++EE G G+ C+EDL+HE+ G HF ++ +
Sbjct 159 ELILKRGQAKVKNKTIP--LTDNTVIEEHLGKFGVICLEDLIHEIAFPGKHFQEISWFLC 216
Query 140 HFQL 143
F L
Sbjct 217 PFHL 220
> 7297909
Length=359
Score = 71.2 bits (173), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 37/117 (31%), Positives = 62/117 (52%), Gaps = 6/117 (5%)
Query 31 RETQEALKALGLPRPYSCTLIANDENATKILRKVSPFVFYGLPSAETIRRLFVTR--ARL 88
+ T + + L + ++ + N + +LR + PFV YG PS +IR L + AR+
Sbjct 111 KTTADIFRTLRMGSRHNAVFMENTKENQLLLRVIEPFVVYGNPSLSSIRELVFKKGFARI 170
Query 89 TSEEGVAPVALSDNMMVEEAFGSLGLFCVEDLVHEVVTCGPHFPQVMQRVGHFQLES 145
++ A+ N MVE+ G G+ C+ED++HE+ T GP+F V + + F L S
Sbjct 171 DGKK----TAIQSNTMVEQQLGDKGVICLEDIIHEICTVGPNFAAVNEFLCAFTLSS 223
> CE11024
Length=244
Score = 70.9 bits (172), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 33/112 (29%), Positives = 60/112 (53%), Gaps = 2/112 (1%)
Query 34 QEALKALGLPRPYSCTLIANDENATKILRKVSPFVFYGLPSAETIRRLFVTRARLTSEEG 93
++AL+ L L + + + ++ +LR + P+V +G P+ +TI L R +
Sbjct 102 RKALQILRLRQINNGVFVKLNKATLPLLRIIEPYVAWGYPNNKTIHDLLYKRGYAKVDGN 161
Query 94 VAPVALSDNMMVEEAFGSLGLFCVEDLVHEVVTCGPHFPQVMQRVGHFQLES 145
P+ +DN +VE++ G + C+EDL HE+ T GPHF + + F+L +
Sbjct 162 RVPI--TDNTIVEQSLGKFNIICLEDLAHEIATVGPHFKEATNFLWPFKLNN 211
> Hs17456110
Length=246
Score = 70.5 bits (171), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 31/114 (27%), Positives = 62/114 (54%), Gaps = 2/114 (1%)
Query 32 ETQEALKALGLPRPYSCTLIANDENATKILRKVSPFVFYGLPSAETIRRLFVTRARLTSE 91
+ ++ L+ L L + ++ T + D+ + +LR V P++ +G P+ +++ L R +
Sbjct 102 KVRKVLQLLRLRQIFNGTFVKLDKASINMLRIVEPYIAWGYPNLKSVNELIYKRG--YGK 159
Query 92 EGVAPVALSDNMMVEEAFGSLGLFCVEDLVHEVVTCGPHFPQVMQRVGHFQLES 145
+AL+DN ++ + G G+ C+EDL+HE+ T G F + + F+L S
Sbjct 160 MNKKRIALTDNTLIARSLGKYGIICMEDLIHEIYTVGKGFKEANNFLWAFKLSS 213
> Hs13649740
Length=198
Score = 69.7 bits (169), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 32/114 (28%), Positives = 62/114 (54%), Gaps = 2/114 (1%)
Query 32 ETQEALKALGLPRPYSCTLIANDENATKILRKVSPFVFYGLPSAETIRRLFVTRARLTSE 91
+ ++ L+ L L R ++ T + ++ + +LR V P+ +G P+ +++ L R +
Sbjct 41 KVRKMLQLLCLRRVFNGTFVKLNKASINMLRIVEPYTAWGCPNLKSVNELIYKRG--YGK 98
Query 92 EGVAPVALSDNMMVEEAFGSLGLFCVEDLVHEVVTCGPHFPQVMQRVGHFQLES 145
+AL+DN ++ + G G+ C+EDL+HE+ T G HF + + F+L S
Sbjct 99 INKKQIALTDNALIARSLGKYGIICMEDLIHEIYTVGKHFKEANNFLWPFKLSS 152
> SPAC3H5.07
Length=250
Score = 68.6 bits (166), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 33/86 (38%), Positives = 49/86 (56%), Gaps = 2/86 (2%)
Query 60 ILRKVSPFVFYGLPSAETIRRLFVTRARLTSEEGVAPVALSDNMMVEEAFGSLGLFCVED 119
+L+ V P+V YG+P+ +T+R L R + P LSDN ++E A G + VED
Sbjct 133 MLQVVEPYVTYGIPNHKTVRELIYKRGFGKVNKQRIP--LSDNAIIEAALGKYSILSVED 190
Query 120 LVHEVVTCGPHFPQVMQRVGHFQLES 145
L+HE+ T GP+F Q + F+L S
Sbjct 191 LIHEIYTVGPNFKQAANFLWPFKLSS 216
> Hs20545549
Length=248
Score = 67.8 bits (164), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 30/114 (26%), Positives = 62/114 (54%), Gaps = 2/114 (1%)
Query 32 ETQEALKALGLPRPYSCTLIANDENATKILRKVSPFVFYGLPSAETIRRLFVTRARLTSE 91
+ ++ L+ L L + ++ T + ++ + +LR V P++ +G P+ +++ L R +
Sbjct 104 KVRKVLQLLRLRQIFNGTFVKLNKASINMLRIVEPYIAWGYPNLKSVNELIYKRG--YGK 161
Query 92 EGVAPVALSDNMMVEEAFGSLGLFCVEDLVHEVVTCGPHFPQVMQRVGHFQLES 145
+AL+DN ++ + G G+ C+EDL+HE+ T G F + + F+L S
Sbjct 162 INKKRIALTDNALIARSLGKYGIICMEDLIHEIYTVGKRFKEANNFLWPFKLSS 215
> Hs15431301
Length=248
Score = 67.8 bits (164), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 30/114 (26%), Positives = 62/114 (54%), Gaps = 2/114 (1%)
Query 32 ETQEALKALGLPRPYSCTLIANDENATKILRKVSPFVFYGLPSAETIRRLFVTRARLTSE 91
+ ++ L+ L L + ++ T + ++ + +LR V P++ +G P+ +++ L R +
Sbjct 104 KVRKVLQLLRLRQIFNGTFVKLNKASINMLRIVEPYIAWGYPNLKSVNELIYKRG--YGK 161
Query 92 EGVAPVALSDNMMVEEAFGSLGLFCVEDLVHEVVTCGPHFPQVMQRVGHFQLES 145
+AL+DN ++ + G G+ C+EDL+HE+ T G F + + F+L S
Sbjct 162 INKKRIALTDNALIARSLGKYGIICMEDLIHEIYTVGKRFKEANNFLWPFKLSS 215
> At1g80750
Length=247
Score = 64.7 bits (156), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 41/143 (28%), Positives = 75/143 (52%), Gaps = 4/143 (2%)
Query 3 RRNRKPKQQRKP-ECRVYCVVRNHRRGGCR-ETQEALKALGLPRPYSCTLIANDENATKI 60
+R ++PK P + + ++R + +T+ L L L ++ ++ +
Sbjct 71 QRVKRPKSSPPPVKSDLVFIIRIQGKNDMHPKTKRILNNLQLKSVFTGVFAKATDSLFQK 130
Query 61 LRKVSPFVFYGLPSAETIRRLFVTRARLTSEEGVAPVALSDNMMVEEAFGSLGLFCVEDL 120
L KV P+V YG P+ ++++ L + T EG PV L+DN ++E+A G + +EDL
Sbjct 131 LLKVQPYVTYGYPNDKSVKDLIYKKG-CTIIEG-NPVPLTDNNIIEQALGEHKILGIEDL 188
Query 121 VHEVVTCGPHFPQVMQRVGHFQL 143
V+E+ G HF +VM+ +G +L
Sbjct 189 VNEIARVGDHFREVMRFLGPLKL 211
> YNL002c
Length=322
Score = 64.7 bits (156), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 36/105 (34%), Positives = 54/105 (51%), Gaps = 7/105 (6%)
Query 34 QEALKALGLPR---PYSCTLIANDENATKILRKVSPFVFYGLPSAETIRRLFVTRARLT- 89
+A K L L R + + +N +L+ ++P+V G PS +IR L R R+
Sbjct 157 NKAFKILSLLRLVETNTGVFVKLTKNVYPLLKVIAPYVVIGKPSLSSIRSLIQKRGRIIY 216
Query 90 -SEEGVAP--VALSDNMMVEEAFGSLGLFCVEDLVHEVVTCGPHF 131
E P + L+DN +VEE G G+ CVED++HE+ T G F
Sbjct 217 KGENEAEPHEIVLNDNNIVEEQLGDHGIICVEDIIHEIATMGESF 261
> Hs20551897
Length=117
Score = 61.6 bits (148), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 28/84 (33%), Positives = 49/84 (58%), Gaps = 2/84 (2%)
Query 60 ILRKVSPFVFYGLPSAETIRRLFVTRARLTSEEGVAPVALSDNMMVEEAFGSLGLFCVED 119
+LR V P+V +G P+ +++ +L + SE + L+DN ++EE G G+ C+ED
Sbjct 1 MLRIVEPYVTFGFPNPKSVWQLILKHGE--SEVKNKTIPLTDNTVIEEHLGKFGIICLED 58
Query 120 LVHEVVTCGPHFPQVMQRVGHFQL 143
L+HE+ + G HF ++ + F L
Sbjct 59 LIHEIASPGKHFQEMPWFLHPFHL 82
> Hs22064401
Length=159
Score = 61.6 bits (148), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 33/127 (25%), Positives = 64/127 (50%), Gaps = 3/127 (2%)
Query 19 YCVVRNHRRGGCRETQEALKALGLPRPYSCTLIANDENATKILRKVSPFVFYGLPSAETI 78
+CV + T+ A L L + +S T + ++ + +LR V P++ +G P+ +++
Sbjct 8 FCVPAEPKLAFVIRTRGANGLLRLYQIFSGTFVKLNKASVNMLRIVEPYITWGNPNLKSV 67
Query 79 RRLFVTRARLTSEEGVAPVALSDNMMVEEAFGSLGLFCVEDLVHEVVTCGPHFPQVMQRV 138
L R + +AL+DN ++ + G G+ C+EDL+H++ T G F + +
Sbjct 68 NELIYKRGYGKMNK---QIALTDNALIALSLGKYGIICMEDLIHDIYTVGKRFKEANNFL 124
Query 139 GHFQLES 145
F+L S
Sbjct 125 WPFKLFS 131
> Hs17438700
Length=285
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 26/99 (26%), Positives = 54/99 (54%), Gaps = 2/99 (2%)
Query 47 SCTLIANDENATKILRKVSPFVFYGLPSAETIRRLFVTRARLTSEEGVAPVALSDNMMVE 106
S T + ++ + +LR V P++ +G P+ +++ L + + +AL+DN ++
Sbjct 139 SGTFVKLNKASINMLRIVDPYIVWGYPNLKSVNELIYKCGYGKINKKI--IALTDNALIA 196
Query 107 EAFGSLGLFCVEDLVHEVVTCGPHFPQVMQRVGHFQLES 145
+ G G+ C+ED+++E+ T G HF + + F+L S
Sbjct 197 PSLGKYGIICMEDMINEIYTVGKHFKEANNFLWPFELSS 235
> Hs18594828
Length=117
Score = 58.2 bits (139), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 26/84 (30%), Positives = 46/84 (54%), Gaps = 2/84 (2%)
Query 60 ILRKVSPFVFYGLPSAETIRRLFVTRARLTSEEGVAPVALSDNMMVEEAFGSLGLFCVED 119
+L V P+V +G P+ +++R L + + P L+D+ ++EE G G+ C+ED
Sbjct 1 MLHIVEPYVTWGFPNLKSVRELIFKHRQAKVKNKTIP--LTDSTVIEEHLGKFGVICLED 58
Query 120 LVHEVVTCGPHFPQVMQRVGHFQL 143
L+HE+ G HF ++ + F L
Sbjct 59 LIHEIAFPGKHFEEISWFLSPFHL 82
> Hs17452755
Length=185
Score = 57.0 bits (136), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 29/120 (24%), Positives = 64/120 (53%), Gaps = 6/120 (5%)
Query 28 GGCRETQEALKALGLPRPYSCTLIANDENATKILRKVSPFVFYGLPSAETIRRLFVTRA- 86
G + ++ L+ L L + ++ T + ++ +T +L V P++ + P+ +++ L R
Sbjct 37 GVSTKVRKVLQLLCLHQIFNGTFVKLNKASTNMLWIVEPYIAWRYPNLKSVNELIYQRGY 96
Query 87 -RLTSEEGVAPVALSDNMMVEEAFGSLGLFCVEDLVHEVVTCGPHFPQVMQRVGHFQLES 145
+++ ++ +AL +N ++ ++ G G C+EDL+HE T G F + + F+L S
Sbjct 97 GKISKKQ----IALIENALIAQSLGKYGTICLEDLIHETYTVGKCFKEANSFLWPFKLSS 152
> Hs18544441
Length=217
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 30/114 (26%), Positives = 57/114 (50%), Gaps = 6/114 (5%)
Query 33 TQEALKALGLPRPYSCTLIANDENATKILRKVSPFVFYGLPSAETIRRLFVT--RARLTS 90
Q + L L + +S + K+L + P V +G P+ +++ L + +A++ +
Sbjct 74 VQRTIVRLCLKKIFSGVSVKVTPQNLKMLHIIEPCVTWGFPNLKSLWELLLKCGQAKVKN 133
Query 91 EEGVAPVALSDNMMVEEAFGSLGLFCVEDLVHEVVTCGPHFPQVMQRVGHFQLE 144
+ + L+DN +EE G G+ C+EDL+HE+ G HF ++ + F L
Sbjct 134 KT----ILLTDNTAIEEHLGKFGVICLEDLIHEIALPGKHFQEISWFLHPFHLS 183
> Hs17486085
Length=243
Score = 54.7 bits (130), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 30/118 (25%), Positives = 58/118 (49%), Gaps = 2/118 (1%)
Query 26 RRGGCRETQEALKALGLPRPYSCTLIANDENATKILRKVSPFVFYGLPSAETIRRLFVTR 85
R G + + L L + +S + K+LR V V +G P+ +++ L + R
Sbjct 93 RIDGVSLLVQTIARLRLKKIFSGVFVKVTPQNLKMLRIVERCVTWGFPNLKSVWELILKR 152
Query 86 ARLTSEEGVAPVALSDNMMVEEAFGSLGLFCVEDLVHEVVTCGPHFPQVMQRVGHFQL 143
+ + P L+DN ++EE G +G+ ++DL++E+ G HF +++ + F L
Sbjct 153 GQSKVKNKTIP--LTDNTVIEEHLGKVGVIFLKDLIYEIAFPGKHFQEILWFLCPFHL 208
> 7297615
Length=252
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 29/115 (25%), Positives = 57/115 (49%), Gaps = 3/115 (2%)
Query 32 ETQEALKALGLPRPYSCTLIANDENATKILRKVSPFVFYGLPSAETIRRLFVTRARLTSE 91
+ ++ L+ L + + I ++ +LR P++ +G P+ +++R L R +
Sbjct 107 KVRKVLQLFRLRQINNGVFIKLNKATINMLRIAEPYITWGYPNLKSVRELIYKRGFVKHN 166
Query 92 EGVAPVALSDNMMVEEAF-GSLGLFCVEDLVHEVVTCGPHFPQVMQRVGHFQLES 145
P+ +DN ++E + + CVEDLVHE+ T GP+F + F+L +
Sbjct 167 RQRVPI--TDNFVIERKLRQAHQIQCVEDLVHEIFTVGPNFKYASNFLWPFKLNT 219
> Hs18599429
Length=203
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 26/95 (27%), Positives = 48/95 (50%), Gaps = 2/95 (2%)
Query 40 LGLPRPYSCTLIANDENATKILRKVSPFVFYGLPSAETIRRLFVTRARLTSEEGVAPVAL 99
L L + +S + + K+L V +V +G P+ +++R L + + P L
Sbjct 110 LRLKKMFSGVFVKVNPQNLKMLLIVETYVTWGFPNLKSVRELTWKHEQAKVKNKTIP--L 167
Query 100 SDNMMVEEAFGSLGLFCVEDLVHEVVTCGPHFPQV 134
+DN ++EE G C+EDL+HE+ G +F ++
Sbjct 168 TDNTVIEEHLEKFGDICLEDLIHEIAFPGKYFQEI 202
> Hs18595820
Length=183
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 24/88 (27%), Positives = 49/88 (55%), Gaps = 6/88 (6%)
Query 60 ILRKVSPFVFYGLPSAETIRRLFVTRA--RLTSEEGVAPVALSDNMMVEEAFGSLGLFCV 117
+LR V P++ G P+ +++ L +++ + +AL+DN+++ + G G+ C+
Sbjct 99 MLRTVEPYIACGYPNLKSVNELIYKHGYGKISKKR----IALTDNVLIARSLGKYGIICM 154
Query 118 EDLVHEVVTCGPHFPQVMQRVGHFQLES 145
EDL++E+ T G F + + F+L S
Sbjct 155 EDLIYEIYTVGKRFKEANNFLWPFKLSS 182
> Hs22059586
Length=142
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 24/70 (34%), Positives = 40/70 (57%), Gaps = 2/70 (2%)
Query 59 KILRKVSPFVFYGLPSAETIRRLFVTRARLTSEEGVAPVALSDNMMVEEAFGSLGLFCVE 118
K+L V P+V G P+ + +R L + + T + P L+DN ++EE G G+ C+E
Sbjct 54 KMLHIVEPYVTRGFPNVKFVRDLILKCRQATVKNKAIP--LTDNTVIEEYLGKFGVICLE 111
Query 119 DLVHEVVTCG 128
+L+HE+ G
Sbjct 112 ELIHEIALSG 121
> Hs17439312
Length=194
Score = 48.1 bits (113), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 31/118 (26%), Positives = 54/118 (45%), Gaps = 12/118 (10%)
Query 63 KVSPFVFYGLPSAETIRRLFVTRARLTSEEGVAPVALSDNMMVEEAFGSLGLFCVEDLVH 122
KV +V +G P+ ++++ L + R + + P SDN + +E G G+ C+EDL H
Sbjct 81 KVETYVTWGFPNLKSVQELILKRVQAKVKNKTIPP--SDNTVTKEHLGRFGVICLEDLRH 138
Query 123 EVVTCGPHFPQVMQRVGHFQLESLKQVEELEAKRMEY-------GYIRNKIDMKVAQL 173
E+ G +F ++ + F L RM++ GY I+ + QL
Sbjct 139 EMAFPGKNFQEISWFLCPFYLS---MAHHATNNRMDFFKDMGSPGYQGEHINQLILQL 193
> Hs18586190
Length=173
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 18/47 (38%), Positives = 29/47 (61%), Gaps = 0/47 (0%)
Query 97 VALSDNMMVEEAFGSLGLFCVEDLVHEVVTCGPHFPQVMQRVGHFQL 143
+ L+DN ++EE G G+ C+EDL+HE+ G HF ++ + F L
Sbjct 103 IPLTDNTVIEEDLGKFGVICLEDLIHEIAFPGKHFQEISWFLRPFHL 149
> Hs22042096
Length=179
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 22/72 (30%), Positives = 40/72 (55%), Gaps = 2/72 (2%)
Query 63 KVSPFVFYGLPSAETIRRLFVTRARLTSEEGVAPVALSDNMMVEEAFGSLGLFCVEDLVH 122
KV +V +G P+ ++++ L + R + + P SDN + +E G G+ C+EDL H
Sbjct 66 KVETYVTWGFPNLKSVQELILKRVQAKVKNKTIPP--SDNTVTKEHLGRFGVICLEDLRH 123
Query 123 EVVTCGPHFPQV 134
E+ G +F ++
Sbjct 124 EMAFPGKNFQEI 135
> Hs22062310
Length=265
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 23/79 (29%), Positives = 43/79 (54%), Gaps = 6/79 (7%)
Query 67 FVFYGLPSAETIRRLFVTRARLTSEEGVAPVALSDNMMVEEAFGSLGLFCVEDLVHEVVT 126
FV G P+ +++ +L R + +AL+DN +++++ G G+ C+EDL+HE+ T
Sbjct 18 FVIRGNPNLKSVNKLIYKHGR--GKINKKRIALTDNTLIDQSHGQYGIICIEDLIHELYT 75
Query 127 CGPHF--PQ--VMQRVGHF 141
PQ + ++ HF
Sbjct 76 VALQLSSPQSGMKKKTTHF 94
> Hs20471558
Length=203
Score = 46.6 bits (109), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 21/87 (24%), Positives = 47/87 (54%), Gaps = 4/87 (4%)
Query 46 YSCTLIANDENATKILRKVSPFVFYGLPSAETIRRLFVTRAR-LTSEEGVAPVALSDNMM 104
++ T + ++ + +LR V P + + P+ +++ + R +++G+ L+DN +
Sbjct 72 FNGTFVKLNKASINMLRIVEPHIAWEYPNLKSVNEVIYKRGYGKINKKGIT---LTDNTL 128
Query 105 VEEAFGSLGLFCVEDLVHEVVTCGPHF 131
+ + G+ C+EDL+HE+ T G F
Sbjct 129 IAVSLNKYGIICMEDLIHEIYTVGKCF 155
> Hs22058429
Length=184
Score = 45.8 bits (107), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 18/91 (19%), Positives = 43/91 (47%), Gaps = 14/91 (15%)
Query 43 PRPYSCTLIANDENATKILRKVSPFVFYGLPSAETIRRLFVTRARLTSEEGVAPVALSDN 102
P+ ++ + + + ++R P++ + P+ I + +A++DN
Sbjct 103 PKIFNGIFVKLSKASINMVRIGEPYISWRYPNLNKIEK--------------KQIAMTDN 148
Query 103 MMVEEAFGSLGLFCVEDLVHEVVTCGPHFPQ 133
++ ++ G + C+EDL+HE+ T G F +
Sbjct 149 TLIAQSLGKYDIICMEDLIHEIYTVGKGFKE 179
> ECU03g0950
Length=239
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 29/112 (25%), Positives = 55/112 (49%), Gaps = 4/112 (3%)
Query 32 ETQEALKALGLPRPYSCTLIANDENATKILRKVSPFVFYGLPSAETIRRLFVTRARLTSE 91
+ ++ L+ L R +C L+ N+++ K+L+ + V +G E +R+L T+ +
Sbjct 96 KVRKVLELFRLKRINTCVLVRNNKSTRKMLQIIKDHVAFGTIGMELLRKLVYTKG--SGR 153
Query 92 EGVAPVALSDNMMVEEAFGSLGLFCVEDLVHEVVTCGPHFPQVMQRVGHFQL 143
G V L+ N +E+ F + C+E+LVH + F +V + F L
Sbjct 154 NGHVRVKLT-NEFIEDMFDG-KIRCIEELVHHIYNGTEMFKKVNSFLYPFHL 203
> Hs22068585
Length=169
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 21/79 (26%), Positives = 38/79 (48%), Gaps = 2/79 (2%)
Query 34 QEALKALGLPRPYSCTLIANDENATKILRKVSPFVFYGLPSAETIRRLFVTRARLTSEEG 93
Q A+ L L + +S + K+LR V P+V +G P+ +++R + + +
Sbjct 59 QTAIARLHLKKIFSGVFVKVTPQNLKMLRIVEPYVIWGFPNLKSVREFILKHGQAKVNKN 118
Query 94 VAPVALSDNMMVEEAFGSL 112
P L+DN ++EE G
Sbjct 119 TIP--LTDNTVIEEHLGKF 135
> Hs17475750
Length=115
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 18/52 (34%), Positives = 26/52 (50%), Gaps = 0/52 (0%)
Query 94 VAPVALSDNMMVEEAFGSLGLFCVEDLVHEVVTCGPHFPQVMQRVGHFQLES 145
V P DN ++ G G+ C EDL+HE+ T G F + + F+L S
Sbjct 63 VEPYIAGDNALIARTLGKYGIICTEDLIHEIYTVGKCFEEANNFLWSFKLSS 114
> Hs18604030
Length=94
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 18/47 (38%), Positives = 28/47 (59%), Gaps = 0/47 (0%)
Query 97 VALSDNMMVEEAFGSLGLFCVEDLVHEVVTCGPHFPQVMQRVGHFQL 143
+ L+DN ++E+ G G+ C+EDL HE+ G HF +V + F L
Sbjct 13 IPLTDNTVIEKHLGKFGVVCLEDLSHEIAVPGKHFQEVSWLLRPFYL 59
> Hs20537431
Length=410
Score = 42.0 bits (97), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 15/37 (40%), Positives = 24/37 (64%), Gaps = 0/37 (0%)
Query 97 VALSDNMMVEEAFGSLGLFCVEDLVHEVVTCGPHFPQ 133
+AL+ N ++ ++ G G+ C EDL+HE+ T G F Q
Sbjct 155 IALTGNTLIAQSLGKYGIICTEDLIHEIYTVGKCFKQ 191
> Hs22057886
Length=147
Score = 41.6 bits (96), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 16/55 (29%), Positives = 29/55 (52%), Gaps = 0/55 (0%)
Query 97 VALSDNMMVEEAFGSLGLFCVEDLVHEVVTCGPHFPQVMQRVGHFQLESLKQVEE 151
+ + DN + + G G+ C EDL+HE+ T G F + + F + S +Q+ +
Sbjct 53 IRIRDNALTARSLGKYGIICREDLIHEIYTVGKCFKEANNFLWPFSIFSTRQLTD 107
> Hs20550426
Length=185
Score = 39.7 bits (91), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 24/106 (22%), Positives = 44/106 (41%), Gaps = 20/106 (18%)
Query 40 LGLPRPYSCTLIANDENATKILRKVSPFVFYGLPSAETIRRLFVTRARLTSEEGVAPVAL 99
L L + ++ T ++++ +L V P++ +G P +++ L R E +
Sbjct 67 LRLRQIFNGTFAKLNKDSINMLMIVKPYIAWGYPKLKSVNELIYKRGYGKINENI----- 121
Query 100 SDNMMVEEAFGSLGLFCVEDLVHEVVTCGPHFPQVMQRVGHFQLES 145
C EDLV+E+ T G HF + + F+L S
Sbjct 122 ---------------ICREDLVNEIYTVGKHFKETNNFMWPFKLSS 152
> Hs17454008
Length=102
Score = 32.7 bits (73), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 15/58 (25%), Positives = 33/58 (56%), Gaps = 0/58 (0%)
Query 28 GGCRETQEALKALGLPRPYSCTLIANDENATKILRKVSPFVFYGLPSAETIRRLFVTR 85
G + Q+ L+ L L + ++ TL+ ++ + +LR V P++ +G P+ +++ L R
Sbjct 37 GVSPKVQKVLQLLRLHQIFNGTLVKINKASFNMLRIVEPYIAWGYPNPKSVNELVYKR 94
> Hs22042125
Length=96
Score = 30.4 bits (67), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 22/96 (22%), Positives = 46/96 (47%), Gaps = 3/96 (3%)
Query 11 QRKPECRVYCVVRNHRRGGCR-ETQEALKALGLPRPYSCTLIANDENATKILRKVSPFVF 69
QR E ++ V+R G + Q+ L+ L L + ++ + + + +LR V P++
Sbjct 2 QRDTEPKLAFVIRIRGINGVNPKVQKVLQLLRLCQIFNGNFVKLNRASVNMLRIVEPYIA 61
Query 70 YGLPSAETIRRLFVTRARLTSEEGVAPVALSDNMMV 105
G P+ +++ L R + +AL+DN ++
Sbjct 62 CGYPNLKSVNELGYKRG--YGKINKKRIALTDNALI 95
> Hs20533158
Length=314
Score = 30.0 bits (66), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 16/61 (26%), Positives = 33/61 (54%), Gaps = 4/61 (6%)
Query 26 RRGGCRET----QEALKALGLPRPYSCTLIANDENATKILRKVSPFVFYGLPSAETIRRL 81
R GG + ++ L+ L L + +S T + ++ + ILR V P++ +G P+ +++ L
Sbjct 220 RIGGISDVSPKIRKVLQLLHLHQIFSGTFMKLNKASFNILRIVGPYITWGYPNLKSVNEL 279
Query 82 F 82
Sbjct 280 I 280
Lambda K H
0.322 0.137 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2634976318
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40