bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0385_orf1
Length=218
Score E
Sequences producing significant alignments: (Bits) Value
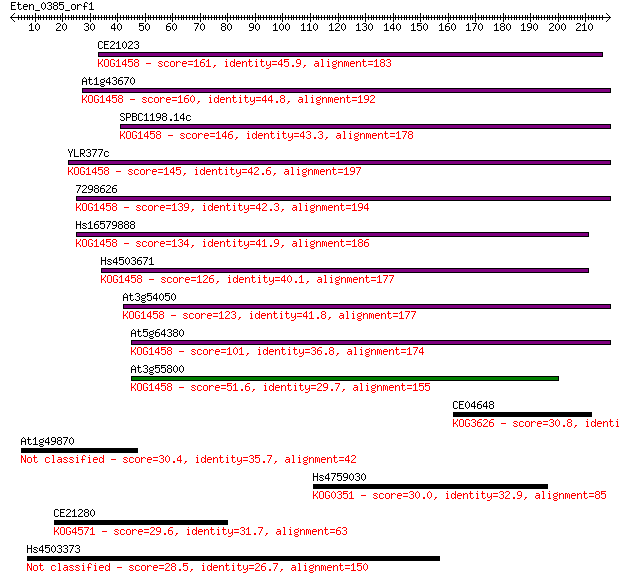
CE21023 161 7e-40
At1g43670 160 2e-39
SPBC1198.14c 146 3e-35
YLR377c 145 7e-35
7298626 139 3e-33
Hs16579888 134 1e-31
Hs4503671 126 3e-29
At3g54050 123 3e-28
At5g64380 101 1e-21
At3g55800 51.6 1e-06
CE04648 30.8 2.3
At1g49870 30.4 2.7
Hs4759030 30.0 3.7
CE21280 29.6 4.9
Hs4503373 28.5 9.8
> CE21023
Length=341
Score = 161 bits (408), Expect = 7e-40, Method: Compositional matrix adjust.
Identities = 84/185 (45%), Positives = 116/185 (62%), Gaps = 4/185 (2%)
Query 33 KDKLKKKGSEAEMNKLLTAIKLAAKVVNREINMAGLVDILGAAGNQNVQGEDQQKLDVFA 92
+++ K + E+ LLT + +A K + AGL + G AG NVQGE+ +KLDV +
Sbjct 23 QEQRKHADASGELTALLTNMLVAIKAIASATQKAGLAKLYGIAGATNVQGEEVKKLDVLS 82
Query 93 NKKFIQALVNREVVCGICTEEDDDFIPVNPNCH--LVLLMDPLDGSSNIDVNVSVGTIFS 150
N+ I L + C + +EE+D+ I V ++ DPLDGSSNID VS+GTIF
Sbjct 83 NELMINMLKSSYTTCLLVSEENDELIEVEEQRRGKYIVTFDPLDGSSNIDCLVSIGTIFG 142
Query 151 IFQRVSPVGQPVERRDFLQPGKEQLAAGYVLYGSSTMLVMTTGSGVNGFTLDSSLGTFFL 210
I+++ P D L+PGKE +AAGY LYGS+TM+V++TG GVNGFTLD S+G F L
Sbjct 143 IYKKRGD--GPATVDDVLKPGKEMVAAGYALYGSATMVVLSTGDGVNGFTLDPSIGEFIL 200
Query 211 SHPNM 215
+HPNM
Sbjct 201 THPNM 205
> At1g43670
Length=341
Score = 160 bits (404), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 86/194 (44%), Positives = 122/194 (62%), Gaps = 6/194 (3%)
Query 27 EFIIANKDKLKKKGSEAEMNKLLTAIKLAAKVVNREINMAGLVDILGAAGNQNVQGEDQQ 86
F++ + K + S + LL+ I L K V +N AGL ++G AG N+QGE+Q+
Sbjct 17 RFVLNEQSKYPE--SRGDFTILLSHIVLGCKFVCSAVNKAGLAKLIGLAGETNIQGEEQK 74
Query 87 KLDVFANKKFIQALVNREVVCGICTEEDDDFIPVNPNCH--LVLLMDPLDGSSNIDVNVS 144
KLDV +N F+ ALV+ + +EED++ V P+ ++ DPLDGSSNID VS
Sbjct 75 KLDVLSNDVFVNALVSSGRTSVLVSEEDEEATFVEPSKRGKYCVVFDPLDGSSNIDCGVS 134
Query 145 VGTIFSIFQRVSPVGQPVERRDFLQPGKEQLAAGYVLYGSSTMLVMTTGSGVNGFTLDSS 204
+GTIF I+ + +P D L+PG E +AAGY +YGSS MLV++TG+GV+GFTLD S
Sbjct 135 IGTIFGIYT-LDHTDEPT-TADVLKPGNEMVAAGYCMYGSSCMLVLSTGTGVHGFTLDPS 192
Query 205 LGTFFLSHPNMHFP 218
LG F L+HP++ P
Sbjct 193 LGEFILTHPDIKIP 206
> SPBC1198.14c
Length=344
Score = 146 bits (369), Expect = 3e-35, Method: Compositional matrix adjust.
Identities = 77/178 (43%), Positives = 113/178 (63%), Gaps = 2/178 (1%)
Query 41 SEAEMNKLLTAIKLAAKVVNREINMAGLVDILGAAGNQNVQGEDQQKLDVFANKKFIQAL 100
+ E++ LL +++ + K + I A LV+++G +G N G++Q+KLD N FI A+
Sbjct 40 ASGELSLLLNSLQFSFKFIANTIRKAELVNLIGLSGIVNSTGDEQKKLDKICNDIFITAM 99
Query 101 VNREVVCGICTEEDDDFIPVNPNCHLVLLMDPLDGSSNIDVNVSVGTIFSIFQRVSPVGQ 160
+ I +EE++D I V+ N + DP+DGSSNID VSVGTIF I+ ++ P G
Sbjct 100 KSNGCCKLIVSEEEEDLIVVDSNGSYAVTCDPIDGSSNIDAGVSVGTIFGIY-KLRP-GS 157
Query 161 PVERRDFLQPGKEQLAAGYVLYGSSTMLVMTTGSGVNGFTLDSSLGTFFLSHPNMHFP 218
+ D L+PGKE +AAGY +YG+S L++TTG VNGFTLD+ +G F L+H NM P
Sbjct 158 QGDISDVLRPGKEMVAAGYTMYGASAHLLLTTGHRVNGFTLDTDIGEFILTHRNMKMP 215
> YLR377c
Length=348
Score = 145 bits (366), Expect = 7e-35, Method: Compositional matrix adjust.
Identities = 84/199 (42%), Positives = 121/199 (60%), Gaps = 7/199 (3%)
Query 22 IPELGEFIIANKDKLKKKGSEAEMNKLLTAIKLAAKVVNREINMAGLVDILGAAGNQNVQ 81
I L FII ++ + K + + +L A++ A K V+ I A LV+++G AG N
Sbjct 20 IITLPRFIIEHQKQFKN--ATGDFTLVLNALQFAFKFVSHTIRRAELVNLVGLAGASNFT 77
Query 82 GEDQQKLDVFANKKFIQALVNREVVCGICTEEDDDFI--PVNPNCHLVLLMDPLDGSSNI 139
G+ Q+KLDV ++ FI A+ ++ + +EE +D I P N + V DP+DGSSN+
Sbjct 78 GDQQKKLDVLGDEIFINAMRASGIIKVLVSEEQEDLIVFPTNTGSYAVCC-DPIDGSSNL 136
Query 140 DVNVSVGTIFSIFQRVSPVGQPVERRDFLQPGKEQLAAGYVLYGSSTMLVMTTGSGVNGF 199
D VSVGTI SIF+ + + D L+ GKE +AA Y +YGSST LV+T G GV+GF
Sbjct 137 DAGVSVGTIASIFRLLPDSSGTIN--DVLRCGKEMVAACYAMYGSSTHLVLTLGDGVDGF 194
Query 200 TLDSSLGTFFLSHPNMHFP 218
TLD++LG F L+HPN+ P
Sbjct 195 TLDTNLGEFILTHPNLRIP 213
> 7298626
Length=334
Score = 139 bits (351), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 82/196 (41%), Positives = 123/196 (62%), Gaps = 6/196 (3%)
Query 25 LGEFIIANKDKLKKKGSEAEMNKLLTAIKLAAKVVNREINMAGLVDILGAAGNQNVQGED 84
L F++ + K K + ++++LL +I+ A K + + AG+ + G AG+ NVQGE+
Sbjct 15 LTRFVLQEQRKFKS--ATGDLSQLLNSIQTAIKATSSAVRKAGIAKLHGFAGDVNVQGEE 72
Query 85 QQKLDVFANKKFIQALVNREVVCGICTEEDDDFIPVN--PNCHLVLLMDPLDGSSNIDVN 142
+KLDV +N+ FI L + C + +EE+++ I V ++ DPLDGSSNID
Sbjct 73 VKKLDVLSNELFINMLKSSYTTCLMVSEENENVIEVEVEKQGKYIVCFDPLDGSSNIDCL 132
Query 143 VSVGTIFSIFQRVSPVGQPVERRDFLQPGKEQLAAGYVLYGSSTMLVMTTGSGVNGFTLD 202
VS+G+IF+I+++ S VE D LQPG + +AAGY LYGS+T +V+ GSGVNGFT D
Sbjct 133 VSIGSIFAIYRKKSDGPPTVE--DALQPGNQLVAAGYALYGSATAIVLGLGSGVNGFTYD 190
Query 203 SSLGTFFLSHPNMHFP 218
++G F L+ PNM P
Sbjct 191 PAIGEFVLTDPNMRVP 206
> Hs16579888
Length=338
Score = 134 bits (338), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 78/188 (41%), Positives = 110/188 (58%), Gaps = 7/188 (3%)
Query 25 LGEFIIANKDKLKKKGSEAEMNKLLTAIKLAAKVVNREINMAGLVDILGAAGNQNVQGED 84
L F++ K + G E+ +LL ++ A K ++ + AG+ + G AG+ NV G+
Sbjct 14 LTRFVMEEGRKARGTG---ELTQLLNSLCTAVKAISSAVRKAGIAHLYGIAGSTNVTGDQ 70
Query 85 QQKLDVFANKKFIQALVNREVVCGICTEEDDDFIPVNPNCH--LVLLMDPLDGSSNIDVN 142
+KLDV +N + L + C + +EED I V P V+ DPLDGSSNID
Sbjct 71 VKKLDVLSNDLVMNMLKSSFATCVLVSEEDKHAIIVEPEKRGKYVVCFDPLDGSSNIDCL 130
Query 143 VSVGTIFSIFQRVSPVGQPVERRDFLQPGKEQLAAGYVLYGSSTMLVMTTGSGVNGFTLD 202
VSVGTIF I+++ S +P E+ D LQPG+ +AAGY LYGS+TMLV+ GVN F LD
Sbjct 131 VSVGTIFGIYRKKS-TDEPSEK-DALQPGRNLVAAGYALYGSATMLVLAMDCGVNCFMLD 188
Query 203 SSLGTFFL 210
++G F L
Sbjct 189 PAIGEFIL 196
> Hs4503671
Length=339
Score = 126 bits (317), Expect = 3e-29, Method: Compositional matrix adjust.
Identities = 71/179 (39%), Positives = 108/179 (60%), Gaps = 4/179 (2%)
Query 34 DKLKKKGSEAEMNKLLTAIKLAAKVVNREINMAGLVDILGAAGNQNVQGEDQQKLDVFAN 93
+K ++ E+ +LL ++ A K ++ + AGL + G AG+ NV G++ +KLDV +N
Sbjct 20 EKGRQAKGTGELTQLLNSMLTAIKAISSAVRKAGLAHLYGIAGSVNVTGDEVKKLDVLSN 79
Query 94 KKFIQALVNREVVCGICTEEDDDFIPV--NPNCHLVLLMDPLDGSSNIDVNVSVGTIFSI 151
I L + C + +EE+ D I V+ DPLDGSSNID S+GTIF+I
Sbjct 80 SLVINMLQSSYSTCVLVSEENKDAIITAKEKRGKYVVCFDPLDGSSNIDCLASIGTIFAI 139
Query 152 FQRVSPVGQPVERRDFLQPGKEQLAAGYVLYGSSTMLVMTTGSGVNGFTLDSSLGTFFL 210
+++ S +P E +D LQ G+ +AAGY LYGS+T++ ++TG GV+ F LD +LG F L
Sbjct 140 YRKTSE-DEPSE-KDALQCGRNIVAAGYALYGSATLVALSTGQGVDLFMLDPALGEFVL 196
> At3g54050
Length=417
Score = 123 bits (308), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 74/194 (38%), Positives = 109/194 (56%), Gaps = 19/194 (9%)
Query 42 EAEMNKLLTAIKLAAKVVNREINMAGLVDILGAAGNQNVQGEDQQKLDVFANKKFIQALV 101
+AE+ ++++I LA K + + AG+ ++ G G N+QGEDQ+KLDV +N+ F L
Sbjct 97 DAELTIVMSSISLACKQIASLVQRAGISNLTGVQGAVNIQGEDQKKLDVISNEVFSNCLR 156
Query 102 NREVVCGICTEEDDDFIPV----NPNCHLVLLMDPLDGSSNIDVNVSVGTIFSIFQ---- 153
+ I +EE+D +PV + + + V++ DPLDGSSNID VS G+IF I+
Sbjct 157 SSGRTGIIASEEED--VPVAVEESYSGNYVVVFDPLDGSSNIDAAVSTGSIFGIYSPNDE 214
Query 154 -------RVSPVGQPVER--RDFLQPGKEQLAAGYVLYGSSTMLVMTTGSGVNGFTLDSS 204
+S +G +R + QPG LAAGY +Y SS + V+T G GV FTLD
Sbjct 215 CIVDDSDDISALGSEEQRCIVNVCQPGNNLLAAGYCMYSSSVIFVLTLGKGVFSFTLDPM 274
Query 205 LGTFFLSHPNMHFP 218
G F L+ N+ P
Sbjct 275 YGEFVLTQENIEIP 288
> At5g64380
Length=404
Score = 101 bits (252), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 64/181 (35%), Positives = 97/181 (53%), Gaps = 12/181 (6%)
Query 45 MNKLLTAIKLAAKVVNREINMAGLVDILGAAGNQNVQGEDQ---QKLDVFANKKFIQALV 101
+ L A K A +V N + LG + G D+ + LD+ +N + +L
Sbjct 94 LYHLQHACKRIASLVASPFNSS-----LGKLSVNSSSGSDRDAPKPLDIVSNDIVLSSLR 148
Query 102 NREVVCGICTEEDDDFIPVNPNCHLVLLMDPLDGSSNIDVNVSVGTIFSIFQRVSPVGQ- 160
N V + +EE+D + + V+++DPLDGS NID ++ GTIF I+ R+ +
Sbjct 149 NSGKVAVMASEENDSPTWIKDDGPYVVVVDPLDGSRNIDASIPTGTIFGIYNRLVELDHL 208
Query 161 PVERR---DFLQPGKEQLAAGYVLYGSSTMLVMTTGSGVNGFTLDSSLGTFFLSHPNMHF 217
PVE + + LQ G +A+GYVLY S+T+ +T GSG + FTLD S G F L+H N+
Sbjct 209 PVEEKAELNSLQRGSRLVASGYVLYSSATIFCVTLGSGTHAFTLDHSTGEFVLTHQNIKI 268
Query 218 P 218
P
Sbjct 269 P 269
> At3g55800
Length=393
Score = 51.6 bits (122), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 46/159 (28%), Positives = 71/159 (44%), Gaps = 23/159 (14%)
Query 45 MNKLLTAIKLAAKVVNREINMAGLVDILGAAGNQNVQGEDQQKLDVFANKKFIQALVNRE 104
+ LL + A + + ++ A G N G++Q +D+ A+K +AL
Sbjct 93 LRTLLMCMGEALRTIAFKVRTASC----GGTACVNSFGDEQLAVDMLADKLLFEALQYSH 148
Query 105 VVCGICTEE----DDDFIPVNPNCHLVLLMDPLDGSSNIDVNVSVGTIFSIFQRVSPVGQ 160
V C+EE D PV + DPLDGSS +D N +VGTIF ++
Sbjct 149 VCKYACSEEVPELQDMGGPVEGGFSVAF--DPLDGSSIVDTNFTVGTIFGVW-------- 198
Query 161 PVERRDFLQPGKEQLAAGYVLYGSSTMLVMTTGSGVNGF 199
P ++ + G +Q+AA +YG T V+ V GF
Sbjct 199 PGDKLTGIT-GGDQVAAAMGIYGPRTTYVL----AVKGF 232
> CE04648
Length=809
Score = 30.8 bits (68), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 21/56 (37%), Positives = 29/56 (51%), Gaps = 6/56 (10%)
Query 162 VERRDFLQPGKEQLAAGYVLYGSSTMLVMTTGSGVNGFTLDSS------LGTFFLS 211
+ER+ ++P K L G V+ + M M TG VN F L+SS +G FLS
Sbjct 498 LERQFSVRPSKANLLIGCVMVPMAGMGCMVTGGIVNHFRLNSSKMLKFAIGLIFLS 553
> At1g49870
Length=828
Score = 30.4 bits (67), Expect = 2.7, Method: Composition-based stats.
Identities = 15/44 (34%), Positives = 30/44 (68%), Gaps = 3/44 (6%)
Query 5 KYIAQKVDVKMPQ--SSAPIPELGEFIIANKDKLKKKGSEAEMN 46
K +++K + K+PQ +S P+P+LG ++ + +L+K+ EA+ N
Sbjct 416 KEVSEK-NKKVPQGVASDPVPDLGSILVKHSSRLEKEIEEAKKN 458
> Hs4759030
Length=1208
Score = 30.0 bits (66), Expect = 3.7, Method: Composition-based stats.
Identities = 28/91 (30%), Positives = 46/91 (50%), Gaps = 10/91 (10%)
Query 111 TEEDDDFIPVNPNCHLVLLMDPLDGSSNIDVNV----SVGTIFSIFQRVSPVGQPVER-- 164
+EED D + P LV P+ ++D V S+G + + + V Q +E+
Sbjct 421 SEEDTDAVGPEP---LVPSPQPVPEVPSLDPTVLPLYSLGPSGQLAETPAEVFQALEQLG 477
Query 165 RDFLQPGKEQLAAGYVLYGSSTMLVMTTGSG 195
+PG+E+ A +L G ST+LV+ TG+G
Sbjct 478 HQAFRPGQER-AVMRILSGISTLLVLPTGAG 507
> CE21280
Length=228
Score = 29.6 bits (65), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 20/69 (28%), Positives = 34/69 (49%), Gaps = 6/69 (8%)
Query 17 QSSAPIPELGEFIIANKDKLKKKGSEAEMNKLLTAIKLAAKVV------NREINMAGLVD 70
Q + PI +L + ++ D LKK+GS + KLL+ + KV ++ L+D
Sbjct 132 QEAIPINDLVDIVMQTVDNLKKEGSSNDETKLLSRKRQQNKVAAARYRDKQKAKWQDLLD 191
Query 71 ILGAAGNQN 79
L A ++N
Sbjct 192 QLEAEEDRN 200
> Hs4503373
Length=1025
Score = 28.5 bits (62), Expect = 9.8, Method: Compositional matrix adjust.
Identities = 40/159 (25%), Positives = 68/159 (42%), Gaps = 29/159 (18%)
Query 7 IAQKVDVKMPQSSAPIPELGEFIIANKDKLKKKGSEAEMNKLLTAIKLAAKVVNREINMA 66
IA+ +D K+P + + + I NK +LK E N + +K + + R I
Sbjct 868 IAELMDKKLPSFGPYLEQRKKIIAENKIRLK------EQNVAFSPLKRSCFIPKRPIPT- 920
Query 67 GLVDILGAAGNQNVQGEDQQKLDVF---ANKKFIQALVNREVV--CGIC--TEEDDDF-- 117
+ D++G A Q L F +N + + A+++ E+ CG C T D +
Sbjct 921 -IKDVIGKA---------LQYLGTFGELSNVEQVVAMIDEEMCINCGKCYMTCNDSGYQA 970
Query 118 IPVNPNCHLVLLMDPLDGSSNIDVNVSVGTIFSIFQRVS 156
I +P HL + D G + + +SV I + VS
Sbjct 971 IQFDPETHLPTITDTCTGCT---LCLSVCPIVDCIKMVS 1006
Lambda K H
0.318 0.136 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4076275884
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40