bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0393_orf1
Length=124
Score E
Sequences producing significant alignments: (Bits) Value
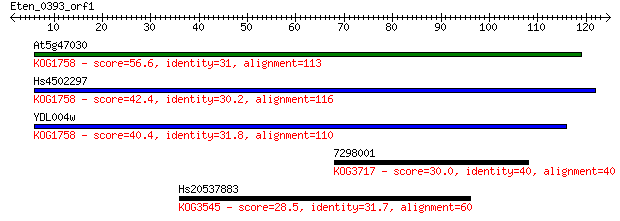
At5g47030 56.6 1e-08
Hs4502297 42.4 2e-04
YDL004w 40.4 8e-04
7298001 30.0 0.95
Hs20537883 28.5 3.4
> At5g47030
Length=203
Score = 56.6 bits (135), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 35/113 (30%), Positives = 58/113 (51%), Gaps = 8/113 (7%)
Query 6 VSLPGSTGHFTVTNGHGPLVTRLRAGPLAVFAANNNNPTTFFVADGFLVLSPPQDDSNCT 65
V +P STG V GH P + L+ G ++V + +F++ GF L +
Sbjct 93 VIIPASTGQMGVLPGHVPTIAELKPGIMSVHEGTDVK--KYFLSSGFAFL------HANS 144
Query 66 LAEVVATEIVPLELLDKEAAAQQLQQLLQEAAAATDQWTKARALLGQELLSAV 118
+A+++A E VPL+ +D + L + Q+ A+AT KA A +G E+ SA+
Sbjct 145 VADIIAVEAVPLDHIDPSQVQKGLAEFQQKLASATTDLEKAEAQIGVEVHSAI 197
> Hs4502297
Length=168
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 35/116 (30%), Positives = 57/116 (49%), Gaps = 7/116 (6%)
Query 6 VSLPGSTGHFTVTNGHGPLVTRLRAGPLAVFAANNNNPTTFFVADGFLVLSPPQDDSNCT 65
V +P TG F + H P + LR G L V A + + +FV+ G + ++ DS+
Sbjct 58 VDVPTLTGAFGILAAHVPTLQVLRPG-LVVVHAEDGTTSKYFVSSGSIAVNA---DSSVQ 113
Query 66 LAEVVATEIVPLELLDKEAAAQQLQQLLQEAAAATDQWTKARALLGQELLSAVIRA 121
L +A E V L++LD AA L++ E D+ T+A + E A+++A
Sbjct 114 L---LAEEAVTLDMLDLGAAKANLEKAQAELVGTADEATRAEIQIRIEANEALVKA 166
> YDL004w
Length=160
Score = 40.4 bits (93), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 35/113 (30%), Positives = 52/113 (46%), Gaps = 15/113 (13%)
Query 6 VSLPGSTGHFTVTNGHGPLVTRLRAGPLAVFAANNNNPTTFFVADGFLVLSPPQDDSNCT 65
V+LP +G V H P V +L G + V +N+ FF++ GF + P D C
Sbjct 52 VNLPAKSGRIGVLANHVPTVEQLLPGVVEVMEGSNSK--KFFISGGFATVQP--DSQLC- 106
Query 66 LAEVVATEIVPLELLDKEAAAQQLQQLLQEA---AAATDQWTKARALLGQELL 115
V A E PLE +E ++ LL EA +++D A A + E+L
Sbjct 107 ---VTAIEAFPLESFSQE----NIKNLLAEAKKNVSSSDAREAAEAAIQVEVL 152
> 7298001
Length=561
Score = 30.0 bits (66), Expect = 0.95, Method: Composition-based stats.
Identities = 16/40 (40%), Positives = 22/40 (55%), Gaps = 2/40 (5%)
Query 68 EVVATEIVPLELLDKEAAAQQLQQLLQEAAAATDQWTKAR 107
E+ + V + +D E A +LQQLL+E A TD W R
Sbjct 71 ELKVEQKVAKKFVDNEGA--KLQQLLEEEAEGTDNWLTPR 108
> Hs20537883
Length=458
Score = 28.5 bits (62), Expect = 3.4, Method: Composition-based stats.
Identities = 19/60 (31%), Positives = 31/60 (51%), Gaps = 3/60 (5%)
Query 36 FAANNNNPTTFFVADGFLVLSPPQDDSNCTLAEVVATEIVPLELLDKEAAAQQLQQLLQE 95
FA + + T +G+ V S QD + VVA E L ++A ++QL+QLL++
Sbjct 16 FAGLDPSKTQISPKEGWQVYSSAQDPDGRCICTVVAPE---QNLCSRDAKSRQLRQLLEK 72
Lambda K H
0.316 0.129 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1187579072
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40