bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0410_orf5
Length=193
Score E
Sequences producing significant alignments: (Bits) Value
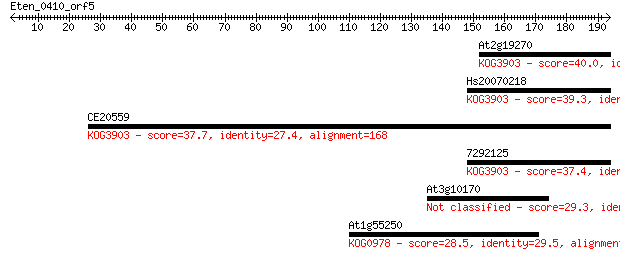
At2g19270 40.0 0.003
Hs20070218 39.3 0.004
CE20559 37.7 0.013
7292125 37.4 0.020
At3g10170 29.3 4.7
At1g55250 28.5 9.0
> At2g19270
Length=359
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 20/42 (47%), Positives = 24/42 (57%), Gaps = 0/42 (0%)
Query 152 MQKRKHQINWLAAEAHDKEMELLELTARTRQTKYQTAMKYGW 193
+ KRKHQI L + KE EL E +R TK +T KYGW
Sbjct 318 LHKRKHQITALFMDMKHKESELTERRSRGLLTKAETQAKYGW 359
> Hs20070218
Length=491
Score = 39.3 bits (90), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 17/46 (36%), Positives = 29/46 (63%), Gaps = 0/46 (0%)
Query 148 EPKVMQKRKHQINWLAAEAHDKEMELLELTARTRQTKYQTAMKYGW 193
+P Q+RKHQI +L +A ++E+EL + + ++ QT KYG+
Sbjct 446 QPTGQQRRKHQITYLIHQAKERELELKNTWSENKLSRRQTQAKYGF 491
> CE20559
Length=405
Score = 37.7 bits (86), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 46/180 (25%), Positives = 74/180 (41%), Gaps = 29/180 (16%)
Query 26 TAEATAEAAAAAAADEAAAERAAAEAAAAEEAA---AAASRYWEYREEDAAVEELGIEKT 82
T ++ + + DE R E EE +AS W +R+ + + K
Sbjct 243 TMNSSMDVVGPSRPDEGMDPRQMYEMPEEEEDVQEGPSASNAWLHRK----INDEQAHKL 298
Query 83 LLEKS-GISTAELRDLRRMGATSVLGATLQNPDWHLTAQAGPQKKAKLRLATKVWSAKDR 141
L+ S I E R + M A S++ D ++ GP K + + + R
Sbjct 299 LMRFSHDIGQEERRSINEM-ANSIV-------DVNVDDALGPDVKTNI-----IKNLGHR 345
Query 142 AITETLE---PKV-----MQKRKHQINWLAAEAHDKEMELLELTARTRQTKYQTAMKYGW 193
A E P+V M +RKHQI +LA+ A +E +L + A +Q+K KYG+
Sbjct 346 AFVEATSAPLPQVQTQGQMSRRKHQITYLASLAVSREEQLKDQWADQKQSKRMARQKYGF 405
> 7292125
Length=472
Score = 37.4 bits (85), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 20/46 (43%), Positives = 27/46 (58%), Gaps = 0/46 (0%)
Query 148 EPKVMQKRKHQINWLAAEAHDKEMELLELTARTRQTKYQTAMKYGW 193
EP +RKHQI +LA +A E EL + + RQT+ T KYG+
Sbjct 427 EPVAGTRRKHQITYLAHKAKANEAELQAMWSANRQTRRATQSKYGF 472
> At3g10170
Length=634
Score = 29.3 bits (64), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 26/39 (66%), Gaps = 0/39 (0%)
Query 135 VWSAKDRAITETLEPKVMQKRKHQINWLAAEAHDKEMEL 173
+WS+K++A+TE +E K+ + QI L+ E +++ EL
Sbjct 411 IWSSKEKALTEAVEEKIRLYKNIQIESLSKEMSEEKKEL 449
> At1g55250
Length=899
Score = 28.5 bits (62), Expect = 9.0, Method: Compositional matrix adjust.
Identities = 18/66 (27%), Positives = 32/66 (48%), Gaps = 5/66 (7%)
Query 110 LQNPDWHLTAQAGPQKKAKLRLATKVWSAKDRAITETLEPKVMQKRKHQI-----NWLAA 164
+Q + HL Q + ++L ++ K T E +VM+K+ HQ+ N+ A
Sbjct 669 MQTQNQHLLQQVAERDDYNIKLVSESVKTKHAYNTHLSEKQVMEKQLHQVNASVENFKAR 728
Query 165 EAHDKE 170
AH++E
Sbjct 729 IAHNEE 734
Lambda K H
0.305 0.115 0.315
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3228275340
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40