bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0414_orf2
Length=169
Score E
Sequences producing significant alignments: (Bits) Value
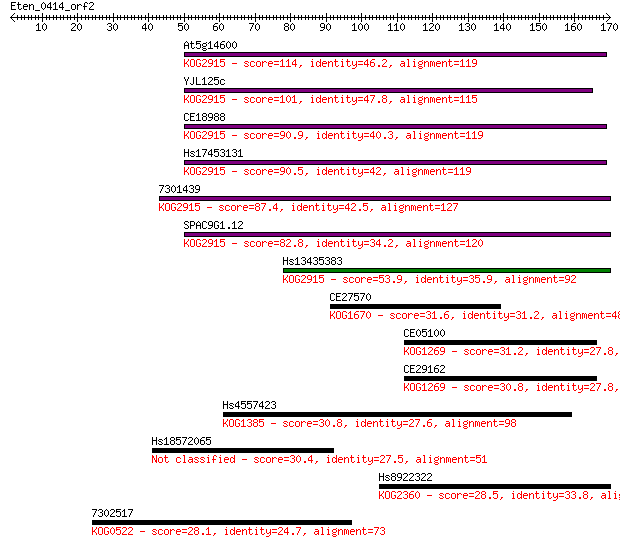
At5g14600 114 7e-26
YJL125c 101 6e-22
CE18988 90.9 1e-18
Hs17453131 90.5 2e-18
7301439 87.4 1e-17
SPAC9G1.12 82.8 3e-16
Hs13435383 53.9 1e-07
CE27570 31.6 0.87
CE05100 31.2 1.1
CE29162 30.8 1.2
Hs4557423 30.8 1.2
Hs18572065 30.4 1.9
Hs8922322 28.5 5.8
7302517 28.1 9.6
> At5g14600
Length=370
Score = 114 bits (286), Expect = 7e-26, Method: Compositional matrix adjust.
Identities = 55/119 (46%), Positives = 83/119 (69%), Gaps = 1/119 (0%)
Query 50 KEGDLVLLYGSRDIIVPCVLRKGGVCRTRLGNFEHADIMRTPMGQKVQDRRSGKWLVVLR 109
++GDLV++Y D++ P + K GV + R G ++H+D + P+G KV + GK++ +L
Sbjct 17 EDGDLVIVYERHDVMKPVKVSKDGVLQNRFGVYKHSDWIGKPLGTKVFSNK-GKFVYLLA 75
Query 110 PTPDLHTLALRHRTQIIYHADISLIMSLLDVCPGKTIIEAGTGSGSLTVSLAASLRPGG 168
PTP+L TL L HRTQI+Y ADIS ++ L+V PG ++E+GTGSGSL+ SLA ++ P G
Sbjct 76 PTPELWTLVLSHRTQILYIADISFVVMYLEVVPGCVVLESGTGSGSLSTSLARAVAPTG 134
> YJL125c
Length=383
Score = 101 bits (252), Expect = 6e-22, Method: Compositional matrix adjust.
Identities = 55/119 (46%), Positives = 73/119 (61%), Gaps = 4/119 (3%)
Query 50 KEGDLVLLYGSRDIIVPCVLRKGGVCRTRLGNFEHADIMRTPMGQKVQDRRSGK----WL 105
KEGDL L++ SRD I P + V TR G+F H DI+ P G ++ R G ++
Sbjct 14 KEGDLTLIWVSRDNIKPVRMHSEEVFNTRYGSFPHKDIIGKPYGSQIAIRTKGSNKFAFV 73
Query 106 VVLRPTPDLHTLALRHRTQIIYHADISLIMSLLDVCPGKTIIEAGTGSGSLTVSLAASL 164
VL+PTP+L TL+L HRTQI+Y D S IM L+ P +IEAGTGSGS + + A S+
Sbjct 74 HVLQPTPELWTLSLPHRTQIVYTPDSSYIMQRLNCSPHSRVIEAGTGSGSFSHAFARSV 132
> CE18988
Length=312
Score = 90.9 bits (224), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 48/119 (40%), Positives = 74/119 (62%), Gaps = 2/119 (1%)
Query 50 KEGDLVLLYGSRDIIVPCVLRKGGVCRTRLGNFEHADIMRTPMGQKVQDRRSGKWLVVLR 109
+EGD V++Y + +VP ++++G + G H I+ GQ++ + ++ +LR
Sbjct 16 QEGDTVIVYVTYGQMVPTIVKRGQTLMMKYGALRHEFIIGKRWGQRLSA--TAGYVYILR 73
Query 110 PTPDLHTLALRHRTQIIYHADISLIMSLLDVCPGKTIIEAGTGSGSLTVSLAASLRPGG 168
PT DL T AL RTQI+Y AD + I+SLLD PG I E+GTGSGSL+ ++A ++ P G
Sbjct 74 PTSDLWTRALPRRTQILYTADCAQILSLLDAKPGSVICESGTGSGSLSHAIALTVAPTG 132
> Hs17453131
Length=289
Score = 90.5 bits (223), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 50/120 (41%), Positives = 74/120 (61%), Gaps = 2/120 (1%)
Query 50 KEGDLVLLYGSRDIIVPCVLRKGGVCRTRLGNFEHA-DIMRTPMGQKVQDRRSGKWLVVL 108
KEGD +L +V +++G +TR G H+ D++ P G KV R G W+ VL
Sbjct 11 KEGDTAILSLGHGAMVAVRVQRGAQTQTRHGVLRHSVDLIGRPFGSKVTCGRGG-WVYVL 69
Query 109 RPTPDLHTLALRHRTQIIYHADISLIMSLLDVCPGKTIIEAGTGSGSLTVSLAASLRPGG 168
PTP+L TL L HRTQI+Y DI+LI +L++ PG + E+GTGSGS++ ++ ++ P G
Sbjct 70 HPTPELWTLNLPHRTQILYSTDIALITMMLELRPGSVVCESGTGSGSVSHAIIRTIAPTG 129
> 7301439
Length=317
Score = 87.4 bits (215), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 54/137 (39%), Positives = 79/137 (57%), Gaps = 12/137 (8%)
Query 43 LYPSNFPKEGDLVLLYGSRDII-----VPCVLRKGG-----VCRTRLGNFEHADIMRTPM 92
L P ++GD+V+LY S + VP ++ K G + +T G+ + +I+
Sbjct 4 LKPKTHIEKGDVVILYLSVSSMHAIEAVPEIVNKKGETIPHIFQTNYGSLKVENIIGVEY 63
Query 93 GQKVQDRRSGKWLVVLRPTPDLHTLALRHRTQIIYHADISLIMSLLDVCPGKTIIEAGTG 152
G KV+ S W VL+PTP+L T L HRTQIIY DIS+I+ L+V PG ++E+GTG
Sbjct 64 GSKVE--LSKGWAHVLQPTPELWTQTLPHRTQIIYTPDISMIIHQLEVRPGAVVVESGTG 121
Query 153 SGSLTVSLAASLRPGGR 169
SGSL+ +L+P G
Sbjct 122 SGSLSHYFLRALKPTGH 138
> SPAC9G1.12
Length=364
Score = 82.8 bits (203), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 41/120 (34%), Positives = 72/120 (60%), Gaps = 0/120 (0%)
Query 50 KEGDLVLLYGSRDIIVPCVLRKGGVCRTRLGNFEHADIMRTPMGQKVQDRRSGKWLVVLR 109
+ GDL + + R+ ++P + + G F H++++ G+++ ++ +L+
Sbjct 11 ENGDLAVAWIGRNKLIPLHIEAEKTFHNQYGAFPHSEMIGKRYGEQIASTAKQGFIYLLQ 70
Query 110 PTPDLHTLALRHRTQIIYHADISLIMSLLDVCPGKTIIEAGTGSGSLTVSLAASLRPGGR 169
PTP+L TLAL HRTQI+Y DI+LI L + G +IEAGTGS S++ +++ ++ P GR
Sbjct 71 PTPELWTLALPHRTQIVYTPDIALIHQKLRITYGTRVIEAGTGSASMSHAISRTVGPLGR 130
> Hs13435383
Length=477
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 33/101 (32%), Positives = 60/101 (59%), Gaps = 9/101 (8%)
Query 78 RLGNFE--HADIMRTPMGQKV-----QDRRS--GKWLVVLRPTPDLHTLALRHRTQIIYH 128
RL NF +++ P G+ V Q RS GK ++ RP + + + ++ T I +
Sbjct 173 RLNNFGLLNSNWGAVPFGKIVGKFPGQILRSSFGKQYMLRRPALEDYVVLMKRGTAITFP 232
Query 129 ADISLIMSLLDVCPGKTIIEAGTGSGSLTVSLAASLRPGGR 169
DI++I+S++D+ PG T++EAG+GSG +++ L+ ++ GR
Sbjct 233 KDINMILSMMDINPGDTVLEAGSGSGGMSLFLSKAVGSQGR 273
> CE27570
Length=251
Score = 31.6 bits (70), Expect = 0.87, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 28/48 (58%), Gaps = 6/48 (12%)
Query 91 PMGQKVQDRRSGKWLVVLRPTPDLHTLALRHRTQIIYHADISLIMSLL 138
PM + V + + G+WLVV + L+ RTQ++ H + L+M+++
Sbjct 95 PMWEDVNNVQGGRWLVV------VDKQKLQRRTQLLDHYWLELLMAIV 136
> CE05100
Length=495
Score = 31.2 bits (69), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 15/54 (27%), Positives = 29/54 (53%), Gaps = 0/54 (0%)
Query 112 PDLHTLALRHRTQIIYHADISLIMSLLDVCPGKTIIEAGTGSGSLTVSLAASLR 165
PD +++ L H + + +D + I++ L + K +++ G G G T LA + R
Sbjct 55 PDTNSMMLNHSAEELESSDRADILASLPLLHNKDVVDIGAGIGRFTTVLAETAR 108
> CE29162
Length=484
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 15/54 (27%), Positives = 29/54 (53%), Gaps = 0/54 (0%)
Query 112 PDLHTLALRHRTQIIYHADISLIMSLLDVCPGKTIIEAGTGSGSLTVSLAASLR 165
PD +++ L H + + +D + I++ L + K +++ G G G T LA + R
Sbjct 44 PDTNSMMLNHSAEELESSDRADILASLPLLHNKDVVDIGAGIGRFTTVLAETAR 97
> Hs4557423
Length=484
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 27/98 (27%), Positives = 42/98 (42%), Gaps = 11/98 (11%)
Query 61 RDIIVPCVLRKGGVCRTRLGNFEHADIMRTPMGQKVQDRRSGKWLVVLRPTPDLHTLALR 120
++++ PC+ + G +EHA++ GQK S L R + L
Sbjct 319 KELVSPCL------SPSFKGEWEHAEVTYRVSGQKAA--ASLHELCAARVSEVLQNRV-- 368
Query 121 HRTQIIYHADISLIMSLLDVCPGKTIIEAGTGSGSLTV 158
HRT+ + H D D+ G +I+A G GSL V
Sbjct 369 HRTEEVKHVDFYAFSYYYDLAAGVGLIDAEKG-GSLVV 405
> Hs18572065
Length=303
Score = 30.4 bits (67), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 14/53 (26%), Positives = 28/53 (52%), Gaps = 2/53 (3%)
Query 41 QDLYPSNFPKEGDLVLLYGSRDI--IVPCVLRKGGVCRTRLGNFEHADIMRTP 91
+DL P+ P + LVL Y + D+ +V L++ C ++ + + A ++ P
Sbjct 217 KDLLPNTVPDQATLVLQYKALDLEGVVTAPLQENEKCTSKDSDMQQASLVLVP 269
> Hs8922322
Length=429
Score = 28.5 bits (62), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 31/66 (46%), Gaps = 1/66 (1%)
Query 105 LVVLRPTPDLHTLALRHRTQIIYHADISLIMS-LLDVCPGKTIIEAGTGSGSLTVSLAAS 163
L+V DLH L +I S + + LLD PG +I+A G+ T LAA
Sbjct 188 LLVFPAQTDLHEHPLYRAGHLILQDRASCLPAMLLDPPPGSHVIDACAAPGNKTSHLAAL 247
Query 164 LRPGGR 169
L+ G+
Sbjct 248 LKNQGK 253
> 7302517
Length=701
Score = 28.1 bits (61), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 18/73 (24%), Positives = 29/73 (39%), Gaps = 4/73 (5%)
Query 24 CIVEESGPDLPEHFTKEQDLYPSNFPKEGDLVLLYGSRDIIVPCVLRKGGVCRTRLGNFE 83
C+V++ D+P H+T P + D +L Y I ++ G C + +
Sbjct 483 CLVDDRCFDIPAHYTNRGSDVRRQIPLDEDDMLQYA----IEQSLVETSGACGVDARDDD 538
Query 84 HADIMRTPMGQKV 96
DI GQ V
Sbjct 539 KVDIWEVLRGQNV 551
Lambda K H
0.321 0.140 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2454498488
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40