bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0415_orf3
Length=249
Score E
Sequences producing significant alignments: (Bits) Value
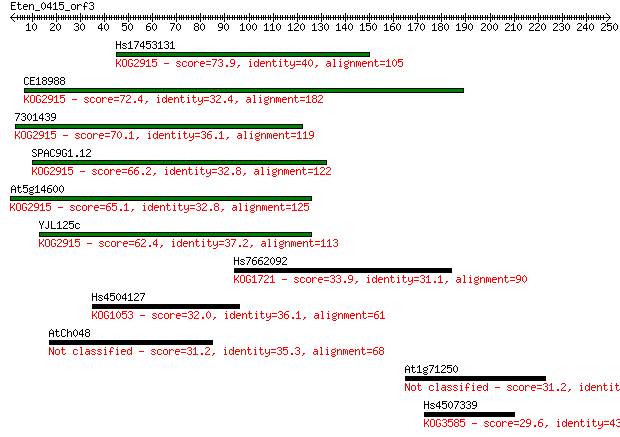
Hs17453131 73.9 3e-13
CE18988 72.4 8e-13
7301439 70.1 5e-12
SPAC9G1.12 66.2 6e-11
At5g14600 65.1 1e-10
YJL125c 62.4 8e-10
Hs7662092 33.9 0.30
Hs4504127 32.0 1.1
AtCh048 31.2 2.0
At1g71250 31.2 2.2
Hs4507339 29.6 5.3
> Hs17453131
Length=289
Score = 73.9 bits (180), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 42/107 (39%), Positives = 54/107 (50%), Gaps = 2/107 (1%)
Query 45 VARAAQQQCGECLPMPHSADGVFLDLPSPWLAIKHASVALK-GGGRLVTFSPCLEQLHKT 103
V Q C + H AD VFLD+PSPW A+ HA ALK GGR +FSPC+EQ+ +T
Sbjct 157 VTVRTQDVCRSGFGVSHVADAVFLDIPSPWEAVGHAWDALKVEGGRFCSFSPCIEQVQRT 216
Query 104 LAAAQEHGFQDFQAFEILAKPWGA-CISRPTPDQRKHEDDVEEGDVS 149
A GF + E+L + + +S P PD D D S
Sbjct 217 CQALAARGFSELSTLEVLPQVYNVRTVSLPPPDLGTGTDGPAGSDTS 263
> CE18988
Length=312
Score = 72.4 bits (176), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 59/192 (30%), Positives = 85/192 (44%), Gaps = 55/192 (28%)
Query 7 EFDLLGISSHVEAIHRDVCADGFCQNVEANTAPAPIAPVARAAQQQCGECLPMPHSADGV 66
EF + G+ S A+ R+VC GF +VE ++ADGV
Sbjct 150 EFKVHGLGSVTTAVVRNVCTSGF--HVE--------------------------NAADGV 181
Query 67 FLDLPSPWLAIKHA--SVALKGGGRLVTFSPCLEQLHKTLAAAQEHGFQDFQAFEILAKP 124
FLD+P+PW AI A S+ GGRLV+FSPCLEQ+ + A GF + E++
Sbjct 182 FLDVPAPWEAIPFAAKSIFRARGGRLVSFSPCLEQVQRACLAMAAAGFVSIETIELV--- 238
Query 125 WGACISRPTPDQRK----HEDDVEE----GDVSHNLKAEKQSERGAQGGQVQQPTPFRDL 176
P+Q+K H +EE GD K+ +R G V+Q +
Sbjct 239 ---------PEQKKIVSQHRQTLEEFEELGDAY-----PKERKRNVDGTIVEQSSSAIPR 284
Query 177 LSHFLQQPSSQP 188
++ + P SQP
Sbjct 285 ITIAIVYPYSQP 296
> 7301439
Length=317
Score = 70.1 bits (170), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 43/120 (35%), Positives = 57/120 (47%), Gaps = 27/120 (22%)
Query 3 EARIEFDLLGISSHVEAIHRDVCADGFCQNVEANTAPAPIAPVARAAQQQCGECLPMPHS 62
+AR EF GI+ V HRDVC GF ++
Sbjct 151 QARDEFRRHGIADFVTVYHRDVCNLGFGAELDGK-------------------------- 184
Query 63 ADGVFLDLPSPWLAIKHASVALK-GGGRLVTFSPCLEQLHKTLAAAQEHGFQDFQAFEIL 121
AD VFLDLP+P LA+ HA ALK GGR +FSPC+EQ + + + GF + + E+L
Sbjct 185 ADAVFLDLPAPDLAVPHAFKALKLSGGRFCSFSPCIEQSQRCIQELTKLGFNEIVSLEVL 244
> SPAC9G1.12
Length=364
Score = 66.2 bits (160), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 40/124 (32%), Positives = 53/124 (42%), Gaps = 29/124 (23%)
Query 10 LLGISSHVEAIHRDVCADGFCQNVEANTAPAPIAPVARAAQQQCGECLPMPHSADGVFLD 69
L+ + + HRDVC DGF L D +FLD
Sbjct 154 LIDVGGNTHLTHRDVCKDGF---------------------------LDTEVKVDAIFLD 186
Query 70 LPSPWLAIKHAS--VALKGGGRLVTFSPCLEQLHKTLAAAQEHGFQDFQAFEILAKPWGA 127
LP+PW AI H S V R+ FSPC+EQ+ + A +E G+ D + E+ K W A
Sbjct 187 LPAPWEAIPHLSNHVNHDKSTRICCFSPCIEQIQHSAEALRELGWCDIEMIEVDYKQWAA 246
Query 128 CISR 131
SR
Sbjct 247 RKSR 250
> At5g14600
Length=370
Score = 65.1 bits (157), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 41/125 (32%), Positives = 61/125 (48%), Gaps = 27/125 (21%)
Query 1 STEARIEFDLLGISSHVEAIHRDVCADGFCQNVEANTAPAPIAPVARAAQQQCGECLPMP 60
+ AR +F+ GISS V RD+ +GF P ++ +
Sbjct 146 AVSAREDFEKTGISSLVTVEVRDIQGEGF---------PEKLSGL--------------- 181
Query 61 HSADGVFLDLPSPWLAIKHASVALKGGGRLVTFSPCLEQLHKTLAAAQEHGFQDFQAFEI 120
AD VFLDLP PWLA+ A+ LK G L +FSPC+EQ+ +T + F + + FE+
Sbjct 182 --ADSVFLDLPQPWLAVPSAAKMLKQDGVLCSFSPCIEQVQRTCEVLRSD-FIEIRTFEV 238
Query 121 LAKPW 125
L + +
Sbjct 239 LLRTY 243
> YJL125c
Length=383
Score = 62.4 bits (150), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 42/117 (35%), Positives = 57/117 (48%), Gaps = 17/117 (14%)
Query 13 ISSHVEAIHRDVCADGFCQNVEANTAPAPIAPVARAAQQQCGECLPMPHSADGVFLDLPS 72
I +V HRDVC GF + +T AA +A+ VFLDLP+
Sbjct 158 IDDNVTITHRDVCQGGFLIK-KGDTTSYEFGNNETAASL----------NANVVFLDLPA 206
Query 73 PWLAIKH----ASVALKGGGRLVTFSPCLEQLHKTLAAAQEHGFQDFQAFEILAKPW 125
PW AI H SV K G L FSPC+EQ+ KTL +++G+ D + EI + +
Sbjct 207 PWDAIPHLDSVISVDEKVG--LCCFSPCIEQVDKTLDVLEKYGWTDVEMVEIQGRQY 261
> Hs7662092
Length=1300
Score = 33.9 bits (76), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 28/115 (24%), Positives = 45/115 (39%), Gaps = 25/115 (21%)
Query 94 SPCLEQLHKTLAAAQEHGFQDFQAFEILAKPWG---ACISRPTPDQRKHEDDVEEGDVSH 150
S C+E+L AA+ +QA++++A+ +S+ P QR HED + V
Sbjct 500 SSCIERLQAAAKAAEMDPVNSYQAWQLMARGMAMEHGFLSKEHPLQRNHEDTLANAGVLF 559
Query 151 NLKAEKQSERGAQGGQVQQPTPF----------------------RDLLSHFLQQ 183
+ + + GA G + + P RD LSH L Q
Sbjct 560 DKEKREYVLVGADGSKQKMPADLVHSTKVGSQRDLPSKLDPLESSRDFLSHGLNQ 614
> Hs4504127
Length=1484
Score = 32.0 bits (71), Expect = 1.1, Method: Composition-based stats.
Identities = 22/61 (36%), Positives = 30/61 (49%), Gaps = 1/61 (1%)
Query 35 ANTAPAPIAPVARAAQQQCGECLPMPHSADGVFLDLPSPWLAIKHASVALKGGGRLVTFS 94
A+T P +P AQ++ L HS D F+DL A+ SV+LK GR + S
Sbjct 1276 ASTTKYPQSPTNSKAQKKNRNKLRRQHSYD-TFVDLQKEEAALAPRSVSLKDKGRFMDGS 1334
Query 95 P 95
P
Sbjct 1335 P 1335
> AtCh048
Length=508
Score = 31.2 bits (69), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 31/71 (43%), Gaps = 5/71 (7%)
Query 17 VEAIHRDVCADGFCQNVEANTAPAPIAPVARAAQQQCGECLPMPHS---ADGVFLDLPSP 73
VE + V G N + + PA + AR AQ GE + + +DGVF P
Sbjct 392 VEQVGVTVEFYGGELNGVSYSDPATVKKYARRAQ--LGEIFELDRATLKSDGVFRSSPRG 449
Query 74 WLAIKHASVAL 84
W HAS AL
Sbjct 450 WFTFGHASFAL 460
> At1g71250
Length=374
Score = 31.2 bits (69), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 23/63 (36%), Positives = 32/63 (50%), Gaps = 12/63 (19%)
Query 165 GQVQQPTPFRDLLSHFLQQPSSQPVSKPTT-GSSPLQ----ATAAAAVDVRRKNTLHISS 219
G+ F DLL+ L+ PS P + PTT G+ LQ A+AAA + L +S
Sbjct 79 GRFSNGLTFIDLLARLLEIPSPPPFADPTTSGNRILQGVNYASAAAGI-------LDVSG 131
Query 220 YHY 222
Y+Y
Sbjct 132 YNY 134
> Hs4507339
Length=435
Score = 29.6 bits (65), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 16/37 (43%), Positives = 22/37 (59%), Gaps = 2/37 (5%)
Query 173 FRDLLSHFLQQPSSQPVSKPTTGSSPLQATAAAAVDV 209
FR +H+ P + PVS P++ SPL AAAA D+
Sbjct 376 FRGSPAHY--TPLTHPVSAPSSSGSPLYEGAAAATDI 410
Lambda K H
0.316 0.131 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5080928138
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40