bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0424_orf4
Length=184
Score E
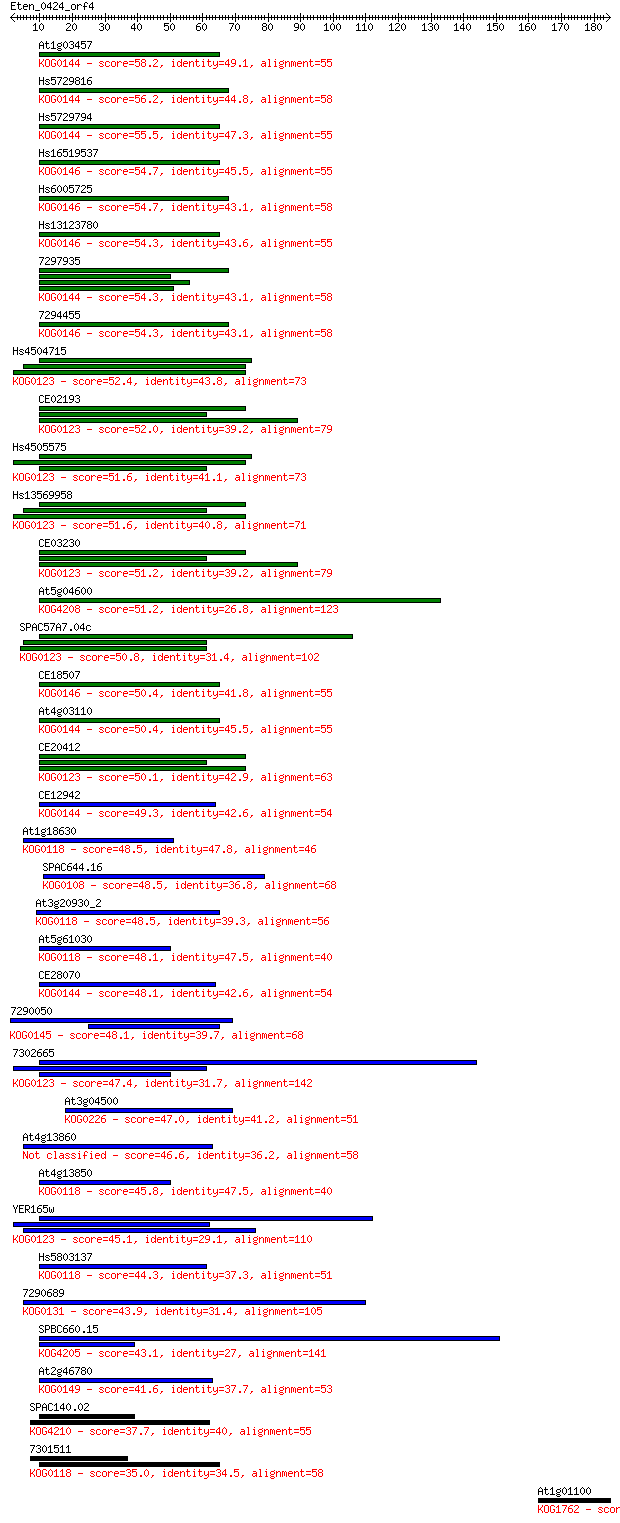
Sequences producing significant alignments: (Bits) Value
At1g03457 58.2 9e-09
Hs5729816 56.2 4e-08
Hs5729794 55.5 6e-08
Hs16519537 54.7 9e-08
Hs6005725 54.7 1e-07
Hs13123780 54.3 1e-07
7297935 54.3 1e-07
7294455 54.3 1e-07
Hs4504715 52.4 6e-07
CE02193 52.0 7e-07
Hs4505575 51.6 8e-07
Hs13569958 51.6 9e-07
CE03230 51.2 1e-06
At5g04600 51.2 1e-06
SPAC57A7.04c 50.8 1e-06
CE18507 50.4 2e-06
At4g03110 50.4 2e-06
CE20412 50.1 2e-06
CE12942 49.3 4e-06
At1g18630 48.5 7e-06
SPAC644.16 48.5 8e-06
At3g20930_2 48.5 8e-06
At5g61030 48.1 8e-06
CE28070 48.1 9e-06
7290050 48.1 1e-05
7302665 47.4 2e-05
At3g04500 47.0 2e-05
At4g13860 46.6 3e-05
At4g13850 45.8 5e-05
YER165w 45.1 8e-05
Hs5803137 44.3 1e-04
7290689 43.9 2e-04
SPBC660.15 43.1 3e-04
At2g46780 41.6 8e-04
SPAC140.02 37.7 0.013
7301511 35.0 0.079
At1g01100 30.0 3.0
> At1g03457
Length=453
Score = 58.2 bits (139), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 27/55 (49%), Positives = 38/55 (69%), Gaps = 0/55 (0%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVSIKK 64
FG V+S V +DK +G +K FGF+SY + AAA A+N MN C +S K+L V +K+
Sbjct 377 FGKVLSAKVFVDKATGISKCFGFISYDSQAAAQNAINTMNGCQLSGKKLKVQLKR 431
> Hs5729816
Length=509
Score = 56.2 bits (134), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 26/58 (44%), Positives = 37/58 (63%), Gaps = 0/58 (0%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVSIKKGEE 67
FG V+S V +DK++ +K FGFVSY P +A AA+ AMN + KRL V +K+ +
Sbjct 447 FGNVISAKVFIDKQTNLSKCFGFVSYDNPVSAQAAIQAMNGFQIGMKRLKVQLKRSKN 504
> Hs5729794
Length=482
Score = 55.5 bits (132), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 26/55 (47%), Positives = 36/55 (65%), Gaps = 0/55 (0%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVSIKK 64
FG VVS V +DK++ +K FGFVSY P +A AA+ +MN + KRL V +K+
Sbjct 420 FGNVVSAKVFIDKQTNLSKCFGFVSYDNPVSAQAAIQSMNGFQIGMKRLKVQLKR 474
> Hs16519537
Length=481
Score = 54.7 bits (130), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 25/55 (45%), Positives = 36/55 (65%), Gaps = 0/55 (0%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVSIKK 64
FGAVVS V +D+ + ++K FGFVS+ P +A A+ AMN + KRL V +K+
Sbjct 419 FGAVVSAKVFVDRATNQSKCFGFVSFDNPTSAQTAIQAMNGFQIGMKRLKVQLKR 473
> Hs6005725
Length=358
Score = 54.7 bits (130), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 25/58 (43%), Positives = 39/58 (67%), Gaps = 0/58 (0%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVSIKKGEE 67
FG V+S V +D+ + ++K FGFVS+ PA+A AA+ AMN + KRL V +K+ ++
Sbjct 296 FGHVISAKVFVDRATNQSKCFGFVSFDNPASAQAAIQAMNGFQIGMKRLKVQLKRPKD 353
> Hs13123780
Length=481
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 24/55 (43%), Positives = 37/55 (67%), Gaps = 0/55 (0%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVSIKK 64
FG ++S V +D+ + ++K FGFVS+ PA+A AA+ AMN + KRL V +K+
Sbjct 423 FGNIISSKVFMDRATNQSKCFGFVSFDNPASAQAAIQAMNGFQIGMKRLKVQLKR 477
> 7297935
Length=1112
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 25/58 (43%), Positives = 39/58 (67%), Gaps = 0/58 (0%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVSIKKGEE 67
FG V+S V +DK++ +K FGFVSY P +A AA+ AM+ + +KRL V +K+ ++
Sbjct 1050 FGNVLSAKVFIDKQTNLSKCFGFVSYDNPHSANAAIQAMHGFQIGSKRLKVQLKRSKD 1107
Score = 45.1 bits (105), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 19/40 (47%), Positives = 27/40 (67%), Gaps = 0/40 (0%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMN 49
FG V+S V +DK++ +K FGFVS+ P +A A+ AMN
Sbjct 766 FGNVISAKVFIDKQTSLSKCFGFVSFDNPDSAQVAIKAMN 805
Score = 35.0 bits (79), Expect = 0.091, Method: Compositional matrix adjust.
Identities = 19/46 (41%), Positives = 28/46 (60%), Gaps = 0/46 (0%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSN 55
+GAV S+ V DK +G +KG FV++ T AA A +A++N N
Sbjct 380 YGAVHSINVLRDKATGISKGCCFVTFYTRHAALKAQDALHNVKTLN 425
Score = 30.4 bits (67), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 16/41 (39%), Positives = 25/41 (60%), Gaps = 0/41 (0%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNN 50
FG V ++ V DK + ++G FV+Y T AA A +A++N
Sbjct 852 FGPVHTLNVLRDKVTSISRGCCFVTYYTRKAALRAQDALHN 892
> 7294455
Length=298
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 25/58 (43%), Positives = 39/58 (67%), Gaps = 0/58 (0%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVSIKKGEE 67
FG V+S V +D+ + ++K FGFVS+ PA+A AA+ AMN + KRL V +K+ ++
Sbjct 236 FGNVISSKVFIDRATNQSKCFGFVSFDNPASAQAAIQAMNGFQIGMKRLKVQLKRPKD 293
> Hs4504715
Length=644
Score = 52.4 bits (124), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 30/66 (45%), Positives = 42/66 (63%), Gaps = 5/66 (7%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVSI-KKGEER 68
FG++ S V L E GR+KGFGFV +++P A AV MN +V +K L V++ ++ EER
Sbjct 317 FGSITSAKVML--EDGRSKGFGFVCFSSPEEATKAVTEMNGRIVGSKPLYVALAQRKEER 374
Query 69 HAAMHL 74
A HL
Sbjct 375 KA--HL 378
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 28/69 (40%), Positives = 36/69 (52%), Gaps = 2/69 (2%)
Query 5 ECLGGFGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVS-IK 63
E FG +SV V D +G++KGFGFVSY A AV MN +S K + V +
Sbjct 209 ELFSQFGKTLSVKVMRD-PNGKSKGFGFVSYEKHEDANKAVEEMNGKEISGKIIFVGRAQ 267
Query 64 KGEERHAAM 72
K ER A +
Sbjct 268 KKVERQAEL 276
Score = 39.3 bits (90), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 24/72 (33%), Positives = 40/72 (55%), Gaps = 3/72 (4%)
Query 2 AAAECLGGFGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVS 61
A + FG ++S V D E+G +KG+ FV + T AA A+ MN L++++++ V
Sbjct 114 ALYDTFSAFGNILSCKVVCD-ENG-SKGYAFVHFETQEAADKAIEKMNGMLLNDRKVFVG 171
Query 62 -IKKGEERHAAM 72
K +ER A +
Sbjct 172 RFKSRKEREAEL 183
> CE02193
Length=692
Score = 52.0 bits (123), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 27/64 (42%), Positives = 41/64 (64%), Gaps = 2/64 (3%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVSI-KKGEER 68
FG + S V +D E+GR+KGFGFV + P A AAV MN+ ++ K L V++ ++ E+R
Sbjct 366 FGTITSAKVMVD-ENGRSKGFGFVCFEKPEEATAAVTDMNSKMIGAKPLYVALAQRKEDR 424
Query 69 HAAM 72
A +
Sbjct 425 RAQL 428
Score = 42.0 bits (97), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 20/51 (39%), Positives = 32/51 (62%), Gaps = 1/51 (1%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNV 60
FG ++S V D E G +KG+GFV + T +A A+ +N L+S+K++ V
Sbjct 168 FGNILSCKVATDDE-GNSKGYGFVHFETEHSAQTAIEKVNGMLLSDKKVYV 217
Score = 39.3 bits (90), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 26/82 (31%), Positives = 41/82 (50%), Gaps = 4/82 (4%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVSI---KKGE 66
FG + S V D + G+ KGFGFV++ AA AV +N+ ++ +S+ +K
Sbjct 261 FGEITSAVVMTDAQ-GKPKGFGFVAFADQDAAGQAVEKLNDSILEGTDCKLSVCRAQKKS 319
Query 67 ERHAAMHLISPAAAQQPTKRQQ 88
ER A + A Q+ +R Q
Sbjct 320 ERSAELKRKYEALKQERVQRYQ 341
> Hs4505575
Length=633
Score = 51.6 bits (122), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 29/66 (43%), Positives = 41/66 (62%), Gaps = 5/66 (7%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVSI-KKGEER 68
FG + S V + E GR+KGFGFV +++P A AV MN +V+ K L V++ ++ EER
Sbjct 314 FGTITSAKVMM--EGGRSKGFGFVCFSSPEEATKAVTEMNGRIVATKPLYVALAQRKEER 371
Query 69 HAAMHL 74
A HL
Sbjct 372 QA--HL 375
Score = 42.4 bits (98), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 25/72 (34%), Positives = 41/72 (56%), Gaps = 3/72 (4%)
Query 2 AAAECLGGFGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVS 61
A + FG ++S V D E+G +KG+GFV + T AA A+ MN L++++++ V
Sbjct 114 ALYDTFSAFGNILSCKVVCD-ENG-SKGYGFVHFETQEAAERAIEKMNGMLLNDRKVFVG 171
Query 62 -IKKGEERHAAM 72
K +ER A +
Sbjct 172 RFKSRKEREAEL 183
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 23/51 (45%), Positives = 32/51 (62%), Gaps = 1/51 (1%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNV 60
FG +SV V D ESG++KGFGFVS+ A AV+ MN ++ K++ V
Sbjct 211 FGPALSVKVMTD-ESGKSKGFGFVSFERHEDAQKAVDEMNGKELNGKQIYV 260
> Hs13569958
Length=631
Score = 51.6 bits (122), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 27/64 (42%), Positives = 40/64 (62%), Gaps = 3/64 (4%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVSI-KKGEER 68
FG + S V + E GR+KGFGFV +++P A AV MN +V+ K L V++ ++ EER
Sbjct 317 FGTITSAKVMM--EGGRSKGFGFVCFSSPEEATKAVTEMNGRIVATKPLYVALAQRKEER 374
Query 69 HAAM 72
A +
Sbjct 375 QAYL 378
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 24/56 (42%), Positives = 34/56 (60%), Gaps = 1/56 (1%)
Query 5 ECLGGFGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNV 60
+ G FG +SV V D ESG++KGFGFVS+ A AV+ MN ++ K++ V
Sbjct 209 DLFGKFGPALSVKVMTD-ESGKSKGFGFVSFERHEDAQKAVDEMNGKELNGKQIYV 263
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/72 (34%), Positives = 41/72 (56%), Gaps = 3/72 (4%)
Query 2 AAAECLGGFGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVS 61
A + + FG ++S V D E+G +KG+GFV + T AA A+ MN L++ +++ V
Sbjct 114 ALYDTVSAFGNILSCNVVCD-ENG-SKGYGFVHFETHEAAERAIKKMNGMLLNGRKVFVG 171
Query 62 -IKKGEERHAAM 72
K +ER A +
Sbjct 172 QFKSRKEREAEL 183
> CE03230
Length=566
Score = 51.2 bits (121), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 27/64 (42%), Positives = 41/64 (64%), Gaps = 2/64 (3%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVSI-KKGEER 68
FG + S V +D E+GR+KGFGFV + P A AAV MN+ ++ K L V++ ++ E+R
Sbjct 366 FGTITSAKVMVD-ENGRSKGFGFVCFEKPEEATAAVTDMNSKMIGAKPLYVALAQRKEDR 424
Query 69 HAAM 72
A +
Sbjct 425 RAQL 428
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 20/51 (39%), Positives = 32/51 (62%), Gaps = 1/51 (1%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNV 60
FG ++S V D E G +KG+GFV + T +A A+ +N L+S+K++ V
Sbjct 168 FGNILSCKVATDDE-GNSKGYGFVHFETEHSAQTAIEKVNGMLLSDKKVYV 217
Score = 38.9 bits (89), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 26/82 (31%), Positives = 41/82 (50%), Gaps = 4/82 (4%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVSI---KKGE 66
FG + S V D + G+ KGFGFV++ AA AV +N+ ++ +S+ +K
Sbjct 261 FGEITSAVVMTDAQ-GKPKGFGFVAFADQDAAGQAVEKLNDSILEGTDCKLSVCRAQKKS 319
Query 67 ERHAAMHLISPAAAQQPTKRQQ 88
ER A + A Q+ +R Q
Sbjct 320 ERSAELKRKYEALKQERVQRYQ 341
> At5g04600
Length=222
Score = 51.2 bits (121), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 33/128 (25%), Positives = 61/128 (47%), Gaps = 11/128 (8%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVSIKKGEERH 69
FG V V V +K++G++K FGF+ + P A A AMN+ L+ L V + + E
Sbjct 83 FGTVKRVRVARNKKTGKSKHFGFIQFEDPEVAEIAAGAMNDYLLMEHMLKVHVIEPENVK 142
Query 70 AAMHL-----ISPAAAQQPTKRQQQQQHQQQQHQQQLQQHMPQSPLPQQQQPQRFNVHAA 124
+ P + Q +RQ ++ ++H++ LQ+ + ++ Q +R + AA
Sbjct 143 PNLWRGFKCNFKPVDSVQIERRQLNKERTLEEHRKMLQK------IVKKDQKRRKRIEAA 196
Query 125 ATPLQQPQ 132
+ P+
Sbjct 197 GIEYECPE 204
> SPAC57A7.04c
Length=653
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 32/96 (33%), Positives = 49/96 (51%), Gaps = 17/96 (17%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVSIKKGEERH 69
FG + S + D E G++KGFGFV YTTP A AV MN +++ K L V++
Sbjct 387 FGTITSAKIMTD-EQGKSKGFGFVCYTTPEEANKAVTEMNQRMLAGKPLYVAL------- 438
Query 70 AAMHLISPAAAQQPTKRQQQQQHQQQQHQQQLQQHM 105
A ++ +R Q + Q ++Q +LQQ +
Sbjct 439 ---------AQRKEVRRSQLEAQIQARNQFRLQQQV 465
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 19/56 (33%), Positives = 34/56 (60%), Gaps = 1/56 (1%)
Query 5 ECLGGFGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNV 60
+ FG ++S V +D E G KG+GFV + + +A AA+ +N L+++K++ V
Sbjct 186 DTFSAFGKILSCKVAVD-ELGNAKGYGFVHFDSVESANAAIEHVNGMLLNDKKVYV 240
Score = 33.9 bits (76), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 18/57 (31%), Positives = 32/57 (56%), Gaps = 1/57 (1%)
Query 4 AECLGGFGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNV 60
++ G FG + S+ + D ++ + +GFGFV+Y A AV+ +N+ K+L V
Sbjct 278 SDLFGQFGEITSLSLVKD-QNDKPRGFGFVNYANHECAQKAVDELNDKEYKGKKLYV 333
> CE18507
Length=459
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 23/55 (41%), Positives = 35/55 (63%), Gaps = 0/55 (0%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVSIKK 64
FG +VS V +D+ + ++K FGFVSY ++ AA+ AMN + KRL V +K+
Sbjct 397 FGHIVSAKVFVDRATNQSKCFGFVSYDNIHSSQAAITAMNGFQIGMKRLKVQLKR 451
> At4g03110
Length=492
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 25/55 (45%), Positives = 36/55 (65%), Gaps = 0/55 (0%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVSIKK 64
FG V+S V +DK +G +K FGFVSY + AAA A++ MN + K+L V +K+
Sbjct 372 FGIVLSAKVFVDKATGVSKCFGFVSYDSQAAAQNAIDMMNGRHLGGKKLKVQLKR 426
> CE20412
Length=646
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 27/64 (42%), Positives = 41/64 (64%), Gaps = 2/64 (3%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVSI-KKGEER 68
+G + S V D E+GR+KGFGFV + P A +AV MN+ +V +K L V+I ++ E+R
Sbjct 340 YGNITSAKVMTD-ENGRSKGFGFVCFEKPEEATSAVTEMNSKMVCSKPLYVAIAQRKEDR 398
Query 69 HAAM 72
A +
Sbjct 399 RAQL 402
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 20/51 (39%), Positives = 32/51 (62%), Gaps = 1/51 (1%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNV 60
FG ++S V +D E G +KG+GFV + T AA A+ +N L++ K++ V
Sbjct 143 FGNILSCKVAID-EDGFSKGYGFVHFETEEAAQNAIQKVNGMLLAGKKVFV 192
Score = 39.3 bits (90), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 36/66 (54%), Gaps = 5/66 (7%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVSI---KKGE 66
FG + S V + G++KGFGFV++ P A AV A+++ + L + + +K
Sbjct 236 FGNITSCEVMTVE--GKSKGFGFVAFANPEEAETAVQALHDSTIEGTDLKLHVCRAQKKS 293
Query 67 ERHAAM 72
ERHA +
Sbjct 294 ERHAEL 299
> CE12942
Length=584
Score = 49.3 bits (116), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 23/54 (42%), Positives = 34/54 (62%), Gaps = 0/54 (0%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVSIK 63
FG ++S V +DK + +K FGFVSY +A A++AMN + +KRL V +K
Sbjct 522 FGGILSAKVFIDKVTNLSKCFGFVSYENAQSATNAISAMNGFQIGSKRLKVQLK 575
> At1g18630
Length=155
Score = 48.5 bits (114), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 22/46 (47%), Positives = 29/46 (63%), Gaps = 0/46 (0%)
Query 5 ECLGGFGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNN 50
E G FG +V V LD+ESG ++GFGFV+Y + A A+ AM N
Sbjct 54 EAFGSFGKIVDAVVVLDRESGLSRGFGFVTYDSIEVANNAMQAMQN 99
> SPAC644.16
Length=422
Score = 48.5 bits (114), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 25/68 (36%), Positives = 36/68 (52%), Gaps = 0/68 (0%)
Query 11 GAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVSIKKGEERHA 70
G V S + +D ESG+ KG+GF Y PA AA+AV +NN +RL V ++
Sbjct 29 GPVKSFQLVIDPESGQPKGYGFCEYHDPATAASAVRNLNNYDAGTRRLRVDFPTADQIRR 88
Query 71 AMHLISPA 78
L+ P+
Sbjct 89 LDKLLGPS 96
> At3g20930_2
Length=92
Score = 48.5 bits (114), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 22/56 (39%), Positives = 34/56 (60%), Gaps = 0/56 (0%)
Query 9 GFGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVSIKK 64
GFG +V V + +DK S R+KG+ F+ YTT AA A+ MN +++ + V + K
Sbjct 22 GFGELVEVKIIMDKISKRSKGYAFLEYTTEEAAGTALKEMNGKIINGWMIVVDVAK 77
> At5g61030
Length=309
Score = 48.1 bits (113), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 19/40 (47%), Positives = 32/40 (80%), Gaps = 0/40 (0%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMN 49
+G VV V LD+E+GR++GFGFV++T+ AA++A+ A++
Sbjct 63 YGEVVDTRVILDRETGRSRGFGFVTFTSSEAASSAIQALD 102
> CE28070
Length=420
Score = 48.1 bits (113), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 23/54 (42%), Positives = 34/54 (62%), Gaps = 0/54 (0%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVSIK 63
FG ++S V +DK + +K FGFVSY +A A++AMN + +KRL V +K
Sbjct 358 FGGILSAKVFIDKVTNLSKCFGFVSYENAQSATNAISAMNGFQIGSKRLKVQLK 411
> 7290050
Length=479
Score = 48.1 bits (113), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 27/68 (39%), Positives = 38/68 (55%), Gaps = 0/68 (0%)
Query 1 AAAAECLGGFGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNV 60
AA + G FGAV SV + D + + KG+GFVS T AA A+ A+N + N+ L V
Sbjct 412 AALWQLFGPFGAVQSVKIVKDPTTNQCKGYGFVSMTNYDEAAMAIRALNGYTMGNRVLQV 471
Query 61 SIKKGEER 68
S K + +
Sbjct 472 SFKTNKAK 479
Score = 35.4 bits (80), Expect = 0.069, Method: Compositional matrix adjust.
Identities = 16/40 (40%), Positives = 24/40 (60%), Gaps = 0/40 (0%)
Query 25 GRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVSIKK 64
G++ G+GFV+Y P A AVN +N + NK + VS +
Sbjct 196 GQSLGYGFVNYVRPQDAEQAVNVLNGLRLQNKTIKVSFAR 235
> 7302665
Length=634
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 44/140 (31%), Positives = 66/140 (47%), Gaps = 20/140 (14%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVSI-KKGEER 68
+G + S V D+E GR+KGFGFV + + A AV +N +V +K L V++ ++ EER
Sbjct 310 YGNITSAKVMTDEE-GRSKGFGFVCFNAASEATCAVTELNGRVVGSKPLYVALAQRKEER 368
Query 69 HAAMHLISPAAAQQPTKRQQQQQHQQQQHQQQL-QQHMPQSP----LPQQQQPQRFNVHA 123
A HL S Q +H QQL Q + P + +P QRF
Sbjct 369 KA--HLAS-----------QYMRHMTGMRMQQLGQIYQPNAASGFFVPTLPSNQRFFGSQ 415
Query 124 AATPLQQPQQLHESLQDPAA 143
AT ++ + ++ PAA
Sbjct 416 VATQMRNTPRWVPQVRPPAA 435
Score = 42.7 bits (99), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 20/40 (50%), Positives = 28/40 (70%), Gaps = 1/40 (2%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMN 49
+G + S Y + KE G++KGFGFV++ T AA AAV A+N
Sbjct 206 YGKITS-YKVMSKEDGKSKGFGFVAFETTEAAEAAVQALN 244
Score = 42.7 bits (99), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 20/59 (33%), Positives = 34/59 (57%), Gaps = 1/59 (1%)
Query 2 AAAECLGGFGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNV 60
A + FG ++S V D E G +KG+GFV + T AA +++ +N L++ K++ V
Sbjct 105 AIYDTFSAFGNILSCKVATD-EKGNSKGYGFVHFETEEAANTSIDKVNGMLLNGKKVYV 162
> At3g04500
Length=245
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 21/51 (41%), Positives = 30/51 (58%), Gaps = 0/51 (0%)
Query 18 VXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVSIKKGEER 68
V DK +G+ KG+GFVS+ PA AAA+ MN V N+ + + +ER
Sbjct 168 VIRDKRTGKTKGYGFVSFLNPADLAAALKEMNGKYVGNRPIKLRKSSWKER 218
> At4g13860
Length=126
Score = 46.6 bits (109), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 21/58 (36%), Positives = 38/58 (65%), Gaps = 0/58 (0%)
Query 5 ECLGGFGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVSI 62
E G+G VV V D+ + R++GFGFV+Y++ + A AAV+ M+ ++ +R++V +
Sbjct 60 EAFSGYGNVVDAIVMRDRYTDRSRGFGFVTYSSHSEAEAAVSGMDGKELNGRRVSVKL 117
> At4g13850
Length=158
Score = 45.8 bits (107), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 19/40 (47%), Positives = 29/40 (72%), Gaps = 0/40 (0%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMN 49
FG VV V +D+E+GR++GFGFV++ AA AA++ M+
Sbjct 58 FGDVVDAKVIVDRETGRSRGFGFVNFNDEGAATAAISEMD 97
> YER165w
Length=577
Score = 45.1 bits (105), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 31/103 (30%), Positives = 50/103 (48%), Gaps = 9/103 (8%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVSI-KKGEER 68
+G + S V + E+G++KGFGFV ++TP A A+ N +V+ K L V+I ++ + R
Sbjct 345 YGTITSAKV-MRTENGKSKGFGFVCFSTPEEATKAITEKNQQIVAGKPLYVAIAQRKDVR 403
Query 69 HAAMHLISPAAAQQPTKRQQQQQHQQQQHQQQLQQHMPQSPLP 111
+ + AQQ R Q + Q MP +P
Sbjct 404 RSQL-------AQQIQARNQMRYQQATAAAAAAAAGMPGQFMP 439
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 20/60 (33%), Positives = 36/60 (60%), Gaps = 1/60 (1%)
Query 2 AAAECLGGFGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVS 61
A + FG ++S + D E+G++KGFGFV + AA A++A+N L++ + + V+
Sbjct 141 ALYDTFSVFGDILSSKIATD-ENGKSKGFGFVHFEEEGAAKEAIDALNGMLLNGQEIYVA 199
Score = 42.7 bits (99), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 41/71 (57%), Gaps = 3/71 (4%)
Query 5 ECLGGFGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVSIKK 64
E FG +VS + D + G+ KGFGFV+Y A AV A+N+ ++ ++L V +
Sbjct 237 ELFAKFGPIVSASLEKDAD-GKLKGFGFVNYEKHEDAVKAVEALNDSELNGEKLYVG--R 293
Query 65 GEERHAAMHLI 75
++++ MH++
Sbjct 294 AQKKNERMHVL 304
> Hs5803137
Length=157
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 19/51 (37%), Positives = 33/51 (64%), Gaps = 0/51 (0%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNV 60
FG + V V D+E+ R++GFGF+++T P A+ A+ AMN + +++ V
Sbjct 29 FGPISEVVVVKDRETQRSRGFGFITFTNPEHASVAMRAMNGESLDGRQIRV 79
> 7290689
Length=347
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 33/114 (28%), Positives = 56/114 (49%), Gaps = 16/114 (14%)
Query 5 ECLGGFGAVVSV-YVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVS-- 61
+ FG ++ + D E+G++K F F+++ + A+ AA++AMN + N+ ++VS
Sbjct 118 DTFSAFGVILQTPKIMRDPETGKSKSFAFINFASFEASDAAMDAMNGQYLCNRPISVSYA 177
Query 62 IKKGE--ERHAAMHLISPAAAQQPTKRQQQQQHQQQQHQ----QQLQQHMPQSP 109
KK ERH +AA++ Q H + HQ +Q MPQ P
Sbjct 178 FKKDHKGERHG-------SAAERLLAAQNPSTHADRPHQLFADAPVQTMMPQMP 224
> SPBC660.15
Length=474
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 38/148 (25%), Positives = 62/148 (41%), Gaps = 35/148 (23%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVSIKKGEERH 69
FG V+ + +DK++GR +GFGFV+Y +A A M+ ++ V +K+
Sbjct 270 FGRVLDATLMMDKDTGRPRGFGFVTYENESAVEA---TMSQPYITIHGKPVEVKR----- 321
Query 70 AAMHLISPAAAQQPTKRQQQQQHQQQQHQQQLQQHMPQSPLPQQQQPQRFNVHAAATP-- 127
A + + R +HQ H + Q N++ TP
Sbjct 322 ---------ATPKASLRDSHDRHQHGYHGNANPYYA-----------QNMNMYGGMTPAM 361
Query 128 ----LQQPQQLHESLQD-PAAATAVQSP 150
+Q QQ E++++ PAAA AV P
Sbjct 362 MAQYYRQMQQYMEAMRNMPAAAGAVPYP 389
Score = 34.7 bits (78), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 12/29 (41%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTP 38
FG V+ V D +GR++GFGF+++ P
Sbjct 186 FGEVLDCTVMRDSTTGRSRGFGFLTFKNP 214
> At2g46780
Length=304
Score = 41.6 bits (96), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 20/53 (37%), Positives = 31/53 (58%), Gaps = 1/53 (1%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVSI 62
FG +V V DK +GR+KG+GFV++ AA A MN ++ +R N ++
Sbjct 45 FGEIVEAVVITDKNTGRSKGYGFVTFKEAEAAMRACQNMNP-VIDGRRANCNL 96
> SPAC140.02
Length=500
Score = 37.7 bits (86), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 13/29 (44%), Positives = 21/29 (72%), Gaps = 0/29 (0%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTP 38
+G +V V +D +SGR+KG+G+V + TP
Sbjct 286 YGTIVGARVIMDGQSGRSKGYGYVDFETP 314
Score = 37.0 bits (84), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 20/57 (35%), Positives = 33/57 (57%), Gaps = 3/57 (5%)
Query 7 LGGFGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNK--RLNVS 61
GG G + S+ + D +SGR KGFG+V+++ +A V MN ++ + RL+ S
Sbjct 386 FGGCGDIQSIRLPTDPQSGRLKGFGYVTFSDIDSAKKCVE-MNGHFIAGRPCRLDFS 441
> 7301511
Length=471
Score = 35.0 bits (79), Expect = 0.079, Method: Compositional matrix adjust.
Identities = 14/30 (46%), Positives = 20/30 (66%), Gaps = 0/30 (0%)
Query 7 LGGFGAVVSVYVXLDKESGRNKGFGFVSYT 36
G +G VV V V D + R++GFGF++YT
Sbjct 52 YGQWGKVVDVVVMRDAATKRSRGFGFITYT 81
Score = 34.7 bits (78), Expect = 0.100, Method: Compositional matrix adjust.
Identities = 19/55 (34%), Positives = 30/55 (54%), Gaps = 3/55 (5%)
Query 10 FGAVVSVYVXLDKESGRNKGFGFVSYTTPAAAAAAVNAMNNCLVSNKRLNVSIKK 64
FG VVSV + DK +G+ +GF FV + A A+ + + K ++V +KK
Sbjct 146 FGNVVSVKLLTDKTTGKRRGFAFVEFDDYDAVDKAILKKQHAI---KYVHVDVKK 197
> At1g01100
Length=112
Score = 30.0 bits (66), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 13/22 (59%), Positives = 15/22 (68%), Gaps = 0/22 (0%)
Query 163 ETKKEDPEEEEEDGDMGLSLFD 184
E KK+D EE DGD+G LFD
Sbjct 91 EEKKKDEPAEESDGDLGFGLFD 112
Lambda K H
0.309 0.122 0.341
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2950576972
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40