bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0428_orf1
Length=200
Score E
Sequences producing significant alignments: (Bits) Value
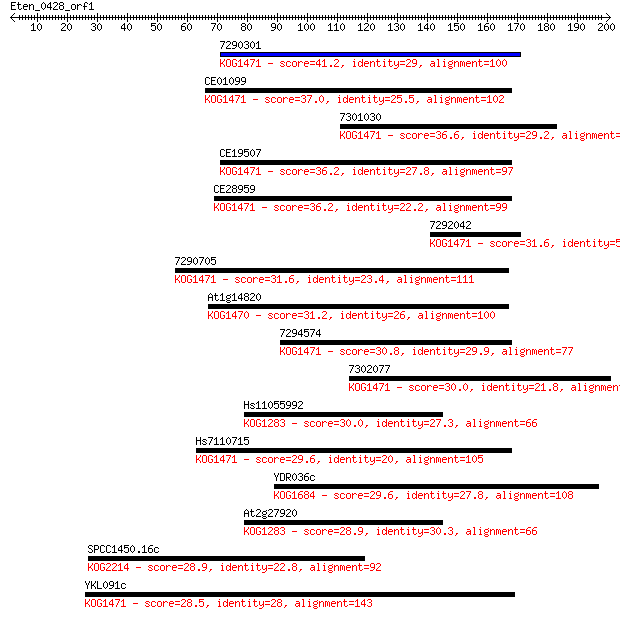
7290301 41.2 0.002
CE01099 37.0 0.028
7301030 36.6 0.036
CE19507 36.2 0.041
CE28959 36.2 0.044
7292042 31.6 1.0
7290705 31.6 1.2
At1g14820 31.2 1.3
7294574 30.8 1.6
7302077 30.0 3.1
Hs11055992 30.0 3.6
Hs7110715 29.6 3.8
YDR036c 29.6 4.6
At2g27920 28.9 6.6
SPCC1450.16c 28.9 7.2
YKL091c 28.5 9.5
> 7290301
Length=303
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 29/101 (28%), Positives = 50/101 (49%), Gaps = 7/101 (6%)
Query 71 VIDAGGFPFRKVVNGSVKMVLDSIVEGLESTVPYVGERTGQVFLINVPSFLNPVIVLVTK 130
++D GG+ R + S+ VL ++ L+ P R + +IN P++L+ +I +++
Sbjct 171 IVDIGGYTLRHLAYVSI-FVLRVYMKFLQEAYP---SRLQAMHVINCPTYLDKLISMMSP 226
Query 131 ILRNGV-NLTSYGSRSKWEPALKRHVGEQHLPREYGGTNST 170
LR V N+ Y + +L + V LP EYGG T
Sbjct 227 FLREEVRNMIRYHTEGM--DSLYKEVPRDMLPNEYGGKAGT 265
> CE01099
Length=743
Score = 37.0 bits (84), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 26/105 (24%), Positives = 49/105 (46%), Gaps = 7/105 (6%)
Query 66 SELTVVIDAGGFPFRKVVNGSVKMVLDSIVEGLESTVPYVGERTGQVFLINVPSFLNPVI 125
S ++V+D G R + V+ +L I+E +E+ P E GQV ++ P +
Sbjct 414 SSWSLVVDLDGLSMRHLWRPGVQCLL-KIIEIVEANYP---ETMGQVLVVRAPRVFPVLW 469
Query 126 VLVTKILRNGVN---LTSYGSRSKWEPALKRHVGEQHLPREYGGT 167
L++ + + S GS + L++H+ E+ +P GG+
Sbjct 470 TLISPFIDEKTRKKFMVSGGSGGDLKEELRKHIEEKFIPDFLGGS 514
> 7301030
Length=353
Score = 36.6 bits (83), Expect = 0.036, Method: Compositional matrix adjust.
Identities = 21/72 (29%), Positives = 38/72 (52%), Gaps = 5/72 (6%)
Query 111 QVFLINVPSFLNPVIVLVTKILRNGVNLTSYGSRSKWEPALKRHVGEQHLPREYGGTNST 170
++++IN+P + ++ + +L VN ++ E L H+G++ LP EYGGTN
Sbjct 241 EMYIINMPKDIQGTVMFLYNVLSMQVNYPIRVLKNSEE--LIEHIGKESLPEEYGGTNG- 297
Query 171 KLHESVVVKYIK 182
H V Y++
Sbjct 298 --HLGECVAYME 307
> CE19507
Length=396
Score = 36.2 bits (82), Expect = 0.041, Method: Compositional matrix adjust.
Identities = 27/105 (25%), Positives = 50/105 (47%), Gaps = 17/105 (16%)
Query 71 VIDAGGFPFR----KVVNGSVKMVLDSIVEGLESTVPYVGERTGQVFLINVPSFLNPVIV 126
++D G F +V G +++ S+ + P E +FLIN PSF+ +
Sbjct 145 ILDLEGLKFDPALISIVTGPYRILWASVY----TAYP---EWINTLFLINAPSFMTLLWK 197
Query 127 LVTKIL----RNGVNLTSYGSRSKWEPALKRHVGEQHLPREYGGT 167
+ +L RN V + S S W+ ++++H ++P+ +GGT
Sbjct 198 AIGPLLPERTRNKVRICS--GNSDWKTSVQKHAHIDNIPKHWGGT 240
> CE28959
Length=408
Score = 36.2 bits (82), Expect = 0.044, Method: Compositional matrix adjust.
Identities = 22/99 (22%), Positives = 49/99 (49%), Gaps = 4/99 (4%)
Query 69 TVVIDAGGFPFRKVVNGSVKMVLDSIVEGLESTVPYVGERTGQVFLINVPSFLNPVIVLV 128
+V+ D G ++ ++K+V +++ L+ P V ++F++N P+F+ + ++
Sbjct 171 SVIFDLDGLSMVQIDLAALKVVT-TMLSQLQEMFPDV---IRKIFIVNTPTFIQVLWSMI 226
Query 129 TKILRNGVNLTSYGSRSKWEPALKRHVGEQHLPREYGGT 167
+ L + W+ LK ++GE+ L +GGT
Sbjct 227 SPCLAKQTQQKVKILGNDWKQHLKENIGEEVLFERWGGT 265
> 7292042
Length=684
Score = 31.6 bits (70), Expect = 1.0, Method: Composition-based stats.
Identities = 15/30 (50%), Positives = 19/30 (63%), Gaps = 1/30 (3%)
Query 141 YGSRSKWEPALKRHVGEQHLPREYGGTNST 170
Y SKWE AL + +Q+LP EYGG N +
Sbjct 601 YVHGSKWE-ALYNQIPKQYLPVEYGGENGS 629
> 7290705
Length=295
Score = 31.6 bits (70), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 26/116 (22%), Positives = 52/116 (44%), Gaps = 7/116 (6%)
Query 56 FWKHVDKRASSELTVVIDAGGFPFRKVVNGSVKMVLDSIVEGLESTVPYVGE----RTGQ 111
F+ D R ++E + G P + +++ + + + L + +V E R +
Sbjct 125 FFMVADCRFATENEERLSDGEIPVFDMAGYTLRHLTKTALGALRVYMKFVQEAHPVRLKE 184
Query 112 VFLINVPSFLNPVIVLVTKILRNGV-NLTSYGSRSKWEPALKRHVGEQHLPREYGG 166
+ ++N PS+++ V+ +V ++ V L + + P RH LP EYGG
Sbjct 185 IHVLNCPSYVDKVMAVVKPFIKGEVFKLIHFHLPNADTPY--RHFPRSMLPEEYGG 238
> At1g14820
Length=252
Score = 31.2 bits (69), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 26/107 (24%), Positives = 44/107 (41%), Gaps = 20/107 (18%)
Query 67 ELTVVIDAGGFPFRKVVNGSVKMVLDS--IVEGLESTVPYVGERTGQVFLINVPSFLNPV 124
+L VID ++ LD+ ++ G + Y ER + +++++P F V
Sbjct 134 KLVAVIDLANITYKN---------LDARGLITGFQFLQSYYPERLAKCYILHMPGFFVTV 184
Query 125 IVLVTKILRNG-----VNLTSYGSRSKWEPALKRHVGEQHLPREYGG 166
V + L V +T + K+E +G LP EYGG
Sbjct 185 WKFVCRFLEKATQEKIVIVTDGEEQRKFE----EEIGADALPEEYGG 227
> 7294574
Length=300
Score = 30.8 bits (68), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 23/77 (29%), Positives = 37/77 (48%), Gaps = 5/77 (6%)
Query 91 LDSIVEGLESTVPYVGERTGQVFLINVPSFLNPVIVLVTKILRNGVNLTSYGSRSKWEPA 150
L SIV+ ++++ P R + +N+P + N +I L +L + L K
Sbjct 169 LRSIVKCIQNSTPM---RHKETHFVNIPHYANRIIELGVSMLSD--KLKKRIIVHKNVDI 223
Query 151 LKRHVGEQHLPREYGGT 167
LK + LP+EYGGT
Sbjct 224 LKTKIDPAILPKEYGGT 240
> 7302077
Length=298
Score = 30.0 bits (66), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 19/88 (21%), Positives = 42/88 (47%), Gaps = 3/88 (3%)
Query 114 LINVPSFLNPVIVLVTKILRNGVNLTSYGSRSKWEPALKRHVGEQHLPREYGGT-NSTKL 172
IN+ F++ ++ L+T ++ LT+ + V ++ LP+EYGG +
Sbjct 188 FINIVPFMDKILALMTPFMKK--ELTTVLHMHSDLKEFYKFVPQEMLPKEYGGQLEEANV 245
Query 173 HESVVVKYIKHSVEKILRRKARHGNNPK 200
+ + K + + ++++ + RH N K
Sbjct 246 AKEIYYKKLLDNRKEMIEFETRHQVNEK 273
> Hs11055992
Length=452
Score = 30.0 bits (66), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 18/66 (27%), Positives = 32/66 (48%), Gaps = 5/66 (7%)
Query 79 FRKVVNGSVKMVLDSIVEGLESTVPYVGERTGQVFLINVPSFLNPVIVLVTKILRNGVNL 138
+++NG ++ L I E G + VF+ F+ PVI +V ++L G+N+
Sbjct 309 LSQLMNGPIRKKLKIIPEDQS-----WGGQATNVFVNMEEDFMKPVISIVDELLEAGINV 363
Query 139 TSYGSR 144
T Y +
Sbjct 364 TVYNGQ 369
> Hs7110715
Length=403
Score = 29.6 bits (65), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 21/105 (20%), Positives = 40/105 (38%), Gaps = 4/105 (3%)
Query 63 RASSELTVVIDAGGFPFRKVVNGSVKMVLDSIVEGLESTVPYVGERTGQVFLINVPSFLN 122
R +T++ D G + + +V+ + + E P E ++F++ P
Sbjct 144 RKVETITIIYDCEGLGLKHLWKPAVEAYGEFLCM-FEENYP---ETLKRLFVVKAPKLFP 199
Query 123 PVIVLVTKILRNGVNLTSYGSRSKWEPALKRHVGEQHLPREYGGT 167
L+ L + W+ L +H+ +P EYGGT
Sbjct 200 VAYNLIKPFLSEDTRKKIMVLGANWKEVLLKHISPDQVPVEYGGT 244
> YDR036c
Length=500
Score = 29.6 bits (65), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 30/119 (25%), Positives = 51/119 (42%), Gaps = 15/119 (12%)
Query 89 MVLDSIVEGLEST---------VPYVGERTGQVFLINVPSFLNPVIVLVTKILRNGVNL- 138
M DS+VE E+T VPY + Q+F+ + S +P L +LRN N+
Sbjct 363 MNQDSLVEFSEATKHKLIDKQRVPYPWTKKEQLFVSQLTSITSPKPSLPMSLLRNTSNVT 422
Query 139 -TSYGSRSKWEPALKRHVGEQHLPREYGGTNSTKLHESVVVKYIKHSVEKILRRKARHG 196
T Y SK++ ++ + ++ + K+ E V + H I R+ + G
Sbjct 423 WTQYPYHSKYQLPTEQEIA-AYIEKRTNDDTGAKVTEREV---LNHFANVIPSRRGKLG 477
> At2g27920
Length=361
Score = 28.9 bits (63), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 20/66 (30%), Positives = 31/66 (46%), Gaps = 5/66 (7%)
Query 79 FRKVVNGSVKMVLDSIVEGLESTVPYVGERTGQVFLINVPSFLNPVIVLVTKILRNGVNL 138
K++NG +K L I L G + VF +F+ PVI V ++L GV++
Sbjct 283 LDKLMNGVIKKKLKIIPNDL-----IWGNNSDDVFTAMEAAFMKPVIEDVDELLATGVDV 337
Query 139 TSYGSR 144
T Y +
Sbjct 338 TIYNGQ 343
> SPCC1450.16c
Length=513
Score = 28.9 bits (63), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 21/92 (22%), Positives = 39/92 (42%), Gaps = 6/92 (6%)
Query 27 CQSGSIVWDVVEKMTHEQLSDFIRYFALFFWKHVDKRASSELTVVIDAGGFPFRKVVNGS 86
C SG+++ ++ E+L+ F WK + + L+ V++ G ++
Sbjct 214 CASGALIASLLSVYRDEELNGLFDTFPSELWKICQQTSDYSLSKVVEYGNMLDISMIASF 273
Query 87 VKMVLDSIVEGLESTVPYVGERTGQVFLINVP 118
V+ L +I T ERTG++ I P
Sbjct 274 VRQRLGTI------TFQEAFERTGRIVNIVAP 299
> YKL091c
Length=310
Score = 28.5 bits (62), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 40/162 (24%), Positives = 69/162 (42%), Gaps = 35/162 (21%)
Query 26 FCQSGSIVWDVVEKMTHEQ--LSDFIRYFALFFWKHV---DKRASSELTV---VIDAGGF 77
F + G I + K+T E+ L + ++ + LF V +RA + V+D G
Sbjct 125 FEELGGINLKKMYKITTEKQMLRNLVKEYELFATYRVPACSRRAGYLIETSCTVLDLKGI 184
Query 78 PFRKVVNGSVKMVLDSIVEGLESTVPYVGERTGQVFLINVP-----------SFLNPVIV 126
+ VL I + + + Y ER G+ ++I+ P FL+PV V
Sbjct 185 SLSNAYH-----VLSYIKDVADISQNYYPERMGKFYIIHSPFGFSTMFKMVKPFLDPVTV 239
Query 127 LVTKILRNGVNLTSYGSRSKWEPALKRHVGEQHLPREYGGTN 168
+KI G S ++ L + + ++LP +YGGT+
Sbjct 240 --SKIFILG---------SSYKKELLKQIPIENLPVKYGGTS 270
Lambda K H
0.322 0.138 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3479363422
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40