bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0437_orf1
Length=176
Score E
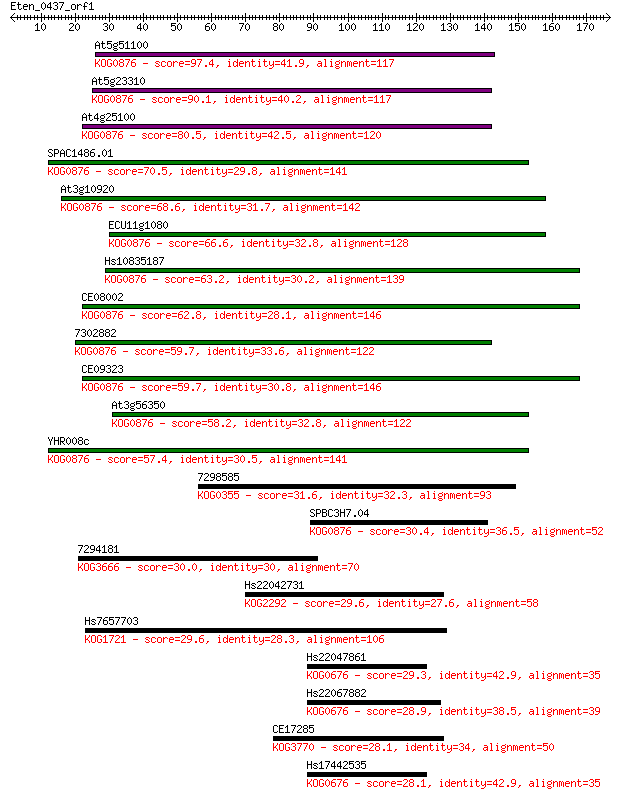
Sequences producing significant alignments: (Bits) Value
At5g51100 97.4 1e-20
At5g23310 90.1 2e-18
At4g25100 80.5 2e-15
SPAC1486.01 70.5 2e-12
At3g10920 68.6 7e-12
ECU11g1080 66.6 2e-11
Hs10835187 63.2 3e-10
CE08002 62.8 4e-10
7302882 59.7 3e-09
CE09323 59.7 3e-09
At3g56350 58.2 1e-08
YHR008c 57.4 1e-08
7298585 31.6 0.97
SPBC3H7.04 30.4 2.0
7294181 30.0 2.8
Hs22042731 29.6 3.2
Hs7657703 29.6 3.6
Hs22047861 29.3 3.7
Hs22067882 28.9 6.2
CE17285 28.1 8.3
Hs17442535 28.1 8.8
> At5g51100
Length=305
Score = 97.4 bits (241), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 49/121 (40%), Positives = 74/121 (61%), Gaps = 6/121 (4%)
Query 26 LSARFSLPPLPYKPDALEPYISAKTIEHHYGKHHKAYVDNLNALVKDSN-NPIHQKLETI 84
++A F L P PY DALEP++S +T+++H+GKHHK YV+NLN + ++ + + + +
Sbjct 49 ITAGFELKPPPYPLDALEPHMSRETLDYHWGKHHKTYVENLNKQILGTDLDALSLEEVVL 108
Query 85 IKNEKGNI---YNQAAQVWNHTFYFNCLGPPSEETAAPLGPTLKRLERDFGSLENFKNEF 141
+ KGN+ +N AAQ WNH F++ + P P G L+ +ERDFGS E F F
Sbjct 109 LSYNKGNMLPAFNNAAQAWNHEFFWESIQPGG--GGKPTGELLRLIERDFGSFEEFLERF 166
Query 142 K 142
K
Sbjct 167 K 167
> At5g23310
Length=263
Score = 90.1 bits (222), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 47/122 (38%), Positives = 72/122 (59%), Gaps = 7/122 (5%)
Query 25 RLSARFSLPPLPYKPDALEPYISAKTIEHHYGKHHKAYVDNLNALVKDSNNPIHQKLETI 84
++ A + L PY DALEPY+S +T+E H+GKHH+ YVDNLN + + +E +
Sbjct 45 KVEAYYGLKTPPYPLDALEPYMSRRTLEVHWGKHHRGYVDNLNKQLGKDDRLYGYTMEEL 104
Query 85 IKNEKGN-----IYNQAAQVWNHTFYFNCLGPPSEETAAPLGPTLKRLERDFGSLENFKN 139
IK N +N AAQV+NH F++ + P +T P L+++++DFGS NF+
Sbjct 105 IKATYNNGNPLPEFNNAAQVYNHDFFWESMQPGGGDT--PQKGVLEQIDKDFGSFTNFRE 162
Query 140 EF 141
+F
Sbjct 163 KF 164
> At4g25100
Length=212
Score = 80.5 bits (197), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 51/125 (40%), Positives = 70/125 (56%), Gaps = 8/125 (6%)
Query 22 AALRLSARFSLPPLPYKPDALEPYISAKTIEHHYGKHHKAYVDNLNALVKDSNNPIHQKL 81
A+ ++A + L P P+ DALEP++S +T+E H+GKHH+AYVDNL V + + L
Sbjct 3 ASSAVTANYVLKPPPFALDALEPHMSKQTLEFHWGKHHRAYVDNLKKQVLGTELE-GKPL 61
Query 82 ETIIKNEKGN-----IYNQAAQVWNHTFYFNCLGPPSEETAAPLGPTLKRLERDFGSLEN 136
E II + N +N AAQ WNH F++ + P + G L LERDF S E
Sbjct 62 EHIIHSTYNNGDLLPAFNNAAQAWNHEFFWESMKPGGGGKPS--GELLALLERDFTSYEK 119
Query 137 FKNEF 141
F EF
Sbjct 120 FYEEF 124
> SPAC1486.01
Length=218
Score = 70.5 bits (171), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 42/145 (28%), Positives = 74/145 (51%), Gaps = 6/145 (4%)
Query 12 TQKTEMALRRAALR--LSARFSLPPLPYKPDALEPYISAKTIEHHYGKHHKAYVDNLNAL 69
++ + A+R ++ + + +LPPLPY +ALEP +S ++ H+ KHH+ YV+NLNA
Sbjct 6 SKNSVAAIRNVSIARGVHTKATLPPLPYAYNALEPALSETIMKLHHDKHHQTYVNNLNAA 65
Query 70 VKDSNNPIHQKLETIIKNEKGNIYNQAAQVWNHTFYFNCLGPPSEETAAPL--GPTLKRL 127
+ +P + LE + + +N + NH+ ++ L P E P+ G K +
Sbjct 66 QEKLADP-NLDLEGEVALQAAIKFNGGGHI-NHSLFWKILAPQKEGGGKPVTSGSLHKAI 123
Query 128 ERDFGSLENFKNEFKKKFSDTSDPG 152
+GSLE+F+ E + G
Sbjct 124 TSKWGSLEDFQKEMNAALASIQGSG 148
> At3g10920
Length=231
Score = 68.6 bits (166), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 45/144 (31%), Positives = 71/144 (49%), Gaps = 6/144 (4%)
Query 16 EMALRRAALRLSARFSLPPLPYKPDALEPYISAKTIEHHYGKHHKAYVDNLNALVKDSNN 75
E + R +R F+LP LPY ALEP IS + ++ H+ KHH+AYV N N ++ +
Sbjct 17 ETSSRLLRIRGIQTFTLPDLPYDYGALEPAISGEIMQIHHQKHHQAYVTNYNNALEQLDQ 76
Query 76 PIHQ-KLETIIKNEKGNIYNQAAQVWNHTFYFNCLGPPSEETAA-PLGPTLKRLERDFGS 133
+++ T++K + +N V NH+ ++ L P SE P G ++ FGS
Sbjct 77 AVNKGDASTVVKLQSAIKFNGGGHV-NHSIFWKNLAPSSEGGGEPPKGSLGSAIDAHFGS 135
Query 134 LENFKNEFKKKFSDTSDPGGCGCC 157
LE KK ++ + G G
Sbjct 136 LEGL---VKKMSAEGAAVQGSGWV 156
> ECU11g1080
Length=233
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 42/139 (30%), Positives = 67/139 (48%), Gaps = 14/139 (10%)
Query 30 FSLPPLPYKPDALEPYISAKTIEHHYGKHHKAYVDNLNALVKDSNNPIHQKLETIIKNEK 89
F LP L Y DALEP I +T+ H+ KHH+AY+++L ++ SN+ + L + +
Sbjct 10 FKLPELDYPYDALEPIIDEETMRTHHSKHHQAYINSLEKTLR-SNSIKGKSLYYYVTKGR 68
Query 90 G----------NIYNQAAQVWNHTFYFNCLGPPSEETAAPLGP-TLKRLERDFGSLENFK 138
G ++ N + +NH+ ++ + PP T+ P+ P L+ +ER FGS E
Sbjct 69 GIRGVKSSARRDLLNFSGGHYNHSLFWKMMCPPG--TSGPMSPRLLEYIERSFGSQEKMV 126
Query 139 NEFKKKFSDTSDPGGCGCC 157
EF G C
Sbjct 127 KEFSNAAVSLFGSGWVWLC 145
> Hs10835187
Length=222
Score = 63.2 bits (152), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 42/140 (30%), Positives = 69/140 (49%), Gaps = 16/140 (11%)
Query 29 RFSLPPLPYKPDALEPYISAKTIEHHYGKHHKAYVDNLNALVKDSNNPIHQKLETI-IKN 87
+ SLP LPY ALEP+I+A+ ++ H+ KHH AYV+NLN + + + T I
Sbjct 25 KHSLPDLPYDYGALEPHINAQIMQLHHSKHHAAYVNNLNVTEEKYQEALAKGDVTAQIAL 84
Query 88 EKGNIYNQAAQVWNHTFYFNCLGPPSEETAAPLGPTLKRLERDFGSLENFKNEFKKKFSD 147
+ +N + NH+ ++ L P P G L+ ++ DFGS + +FK+K +
Sbjct 85 QPALKFNGGGHI-NHSIFWTNLSPNG--GGEPKGELLEAIKLDFGSFD----KFKEKLTA 137
Query 148 TSDPGGCGCCTTLRRTSWSW 167
S ++ + W W
Sbjct 138 AS--------VGVQGSGWGW 149
> CE08002
Length=218
Score = 62.8 bits (151), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 41/147 (27%), Positives = 72/147 (48%), Gaps = 17/147 (11%)
Query 22 AALRLSARFSLPPLPYKPDALEPYISAKTIEHHYGKHHKAYVDNLNALVKDSNNPIHQ-K 80
L + ++ +LP LP+ LEP IS + ++ H+ KHH YV+NLN + + + + +
Sbjct 18 GVLAVRSKHTLPDLPFDYADLEPVISHEIMQLHHQKHHATYVNNLNQIEEKLHEAVSKGN 77
Query 81 LETIIKNEKGNIYNQAAQVWNHTFYFNCLGPPSEETAAPLGPTLKRLERDFGSLENFKNE 140
L+ I + +N + NH+ ++ L E + L T+K RDFGSL+N
Sbjct 78 LKEAIALQPALKFNGGGHI-NHSIFWTNLAKDGGEPSKELMDTIK---RDFGSLDN---- 129
Query 141 FKKKFSDTSDPGGCGCCTTLRRTSWSW 167
+K+ SD + ++ + W W
Sbjct 130 LQKRLSDIT--------IAVQGSGWGW 148
> 7302882
Length=217
Score = 59.7 bits (143), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 41/124 (33%), Positives = 64/124 (51%), Gaps = 7/124 (5%)
Query 20 RRAALRLSARFSLPPLPYKPDALEPYISAKTIEHHYGKHHKAYVDNLNALVKDSNNPIHQ 79
+ A+L + + +LP LPY ALEP I + +E H+ KHH+ YV+NLNA ++
Sbjct 9 QTASLAVRGKHTLPKLPYDYAALEPIICREIMELHHQKHHQTYVNNLNA-AEEQLEEAKS 67
Query 80 KLET--IIKNEKGNIYNQAAQVWNHTFYFNCLGPPSEETAAPLGPTLKRLERDFGSLENF 137
K +T +I+ +N + NHT ++ L P + + L K +E + SLE F
Sbjct 68 KSDTTKLIQLAPALRFNGGGHI-NHTIFWQNLSPNKTQPSDDLK---KAIESQWKSLEEF 123
Query 138 KNEF 141
K E
Sbjct 124 KKEL 127
> CE09323
Length=221
Score = 59.7 bits (143), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 45/157 (28%), Positives = 70/157 (44%), Gaps = 39/157 (24%)
Query 22 AALRLSARFSLPPLPYKPDALEPYISAKTIEHHYGKHHKAYVDNLNALVKDSNNPIHQKL 81
AA+R ++ SLP LPY LEP IS + ++ H+ KHH YV+NLN I +KL
Sbjct 20 AAVR--SKHSLPDLPYDYADLEPVISHEIMQLHHQKHHATYVNNLNQ--------IEEKL 69
Query 82 ETIIKNEKGNI-----------YNQAAQVWNHTFYFNCLGPPSEETAAPLGPTLKRLERD 130
+ KGN+ +N + NH+ ++ L E +A L L ++ D
Sbjct 70 HEAV--SKGNVKEAIALQPALKFNGGGHI-NHSIFWTNLAKDGGEPSAEL---LTAIKSD 123
Query 131 FGSLENFKNEFKKKFSDTSDPGGCGCCTTLRRTSWSW 167
FGSL+N + + ++ + W W
Sbjct 124 FGSLDNLQKQLS------------ASTVAVQGSGWGW 148
> At3g56350
Length=241
Score = 58.2 bits (139), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 40/130 (30%), Positives = 61/130 (46%), Gaps = 15/130 (11%)
Query 31 SLPPLPYKPDALEPYISAKTIEHHYGKHHKAYV-------DNLNALVKDSNNPIHQKLET 83
SLP LPY DALEP IS + + H+ KHH+ YV ++L + + D ++ KL++
Sbjct 37 SLPDLPYAYDALEPAISEEIMRLHHQKHHQTYVTQYNKALNSLRSAMADGDHSSVVKLQS 96
Query 84 IIKNEKGNIYNQAAQVWNHTFYFNCLGPPSEETAA-PLGPTLKRLERDFGSLENFKNEFK 142
+IK +N V NH ++ L P E P P ++ FGSLE +
Sbjct 97 LIK------FNGGGHV-NHAIFWKNLAPVHEGGGKPPHDPLASAIDAHFGSLEGLIQKMN 149
Query 143 KKFSDTSDPG 152
+ + G
Sbjct 150 AEGAAVQGSG 159
> YHR008c
Length=233
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 43/154 (27%), Positives = 64/154 (41%), Gaps = 19/154 (12%)
Query 12 TQKTEMALRRAALRLSARFSLPPLPYKPDALEPYISAKTIEHHYGKHHKAYVDNLNALVK 71
T+K ++L R + + +LP L + ALEPYIS + E HY KHH+ YV+ N V
Sbjct 11 TKKGGLSLLSTTARRT-KVTLPDLKWDFGALEPYISGQINELHYTKHHQTYVNGFNTAVD 69
Query 72 DSNNPIHQKLETIIKNE------------KGNIYNQAAQVWNHTFYFNCLGPPSEETAA- 118
Q+L ++ E + NI NH ++ L P S+
Sbjct 70 Q-----FQELSDLLAKEPSPANARKMIAIQQNIKFHGGGFTNHCLFWENLAPESQGGGEP 124
Query 119 PLGPTLKRLERDFGSLENFKNEFKKKFSDTSDPG 152
P G K ++ FGSL+ K + G
Sbjct 125 PTGALAKAIDEQFGSLDELIKLTNTKLAGVQGSG 158
> 7298585
Length=1423
Score = 31.6 bits (70), Expect = 0.97, Method: Composition-based stats.
Identities = 30/95 (31%), Positives = 42/95 (44%), Gaps = 14/95 (14%)
Query 56 GKHHKAYVDNLNALVKDSNNPIHQKLETIIKNEKGNIYNQAAQVWNHTFYF-NCL-GPPS 113
G+H VDNL I Q LE + K KG I + QV NH + F NCL P+
Sbjct 280 GRHVDHVVDNL----------IKQLLEVLKKKNKGGINIKPFQVRNHLWVFVNCLIENPT 329
Query 114 EETAAPLGPTLKRLERDFGSLENFKNEFKKKFSDT 148
++ TL+ ++ FGS +F S +
Sbjct 330 FDSQTKENMTLQ--QKGFGSKCTLSEKFINNMSKS 362
> SPBC3H7.04
Length=220
Score = 30.4 bits (67), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 19/55 (34%), Positives = 32/55 (58%), Gaps = 3/55 (5%)
Query 89 KGNIYNQAAQVWNHTFYFNCLGPPSEETA-APLGPT-LK-RLERDFGSLENFKNE 140
+ N++N ++Q+ NH F+F+ L P +A A LG LK ++ FGS K++
Sbjct 65 RANLFNYSSQLVNHDFFFSGLISPERPSADADLGAINLKPGIDASFGSFGELKSQ 119
> 7294181
Length=1191
Score = 30.0 bits (66), Expect = 2.8, Method: Composition-based stats.
Identities = 21/70 (30%), Positives = 33/70 (47%), Gaps = 20/70 (28%)
Query 21 RAALRLSARFSLPPLPYKPDALEPYISAKTIEHHYGKHHKAYVDNLNALVKDSNNPIHQK 80
RA L L RF Y++ ++I HHY + Y++ LN + I Q
Sbjct 84 RAQLPLITRF--------------YLAFQSI-HHYASDLQQYIEELN-----TGYYIQQT 123
Query 81 LETIIKNEKG 90
LET+++ E+G
Sbjct 124 LETVLQEEEG 133
> Hs22042731
Length=826
Score = 29.6 bits (65), Expect = 3.2, Method: Composition-based stats.
Identities = 16/58 (27%), Positives = 27/58 (46%), Gaps = 7/58 (12%)
Query 70 VKDSNNPIHQKLETIIKNEKGNIYNQAAQVWNHTFYFNCLGPPSEETAAPLGPTLKRL 127
+K N P+ + K +GN+Y++A +V H E+T LGP +K +
Sbjct 488 MKRENPPVEDSSDEDDKRNQGNLYDKAGKVRKH-------ATEQEKTEEGLGPNIKSI 538
> Hs7657703
Length=1053
Score = 29.6 bits (65), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 30/114 (26%), Positives = 40/114 (35%), Gaps = 24/114 (21%)
Query 23 ALRLSARFSLPPLPYK-PDALEPYISAKTIEHHYGKHHKAYVDNLNALVKDSNNPIHQKL 81
+L+ R PY P E YI A+T+ H K H+ YV L KD+
Sbjct 900 SLKRHVRTHTGERPYVCPVCSEAYIDARTLRKHMTKFHRDYVPCKIMLEKDTL------- 952
Query 82 ETIIKNEKGNIYNQAAQVWNHTFYFNC-------LGPPSEETAAPLGPTLKRLE 128
+NQ QV + GP ET G T++ LE
Sbjct 953 ---------QFHNQGTQVAHAVSILTAGMQEQESSGPQELETVVVTGETMEALE 997
> Hs22047861
Length=660
Score = 29.3 bits (64), Expect = 3.7, Method: Composition-based stats.
Identities = 15/39 (38%), Positives = 21/39 (53%), Gaps = 4/39 (10%)
Query 88 EKGNIYNQAAQVWNHTFYFN-CLGP---PSEETAAPLGP 122
E G ++ ++W+HTFY C+ P P T APL P
Sbjct 66 EHGTNWDDMEKIWHHTFYNKLCMTPEEHPMLLTQAPLNP 104
> Hs22067882
Length=214
Score = 28.9 bits (63), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 15/41 (36%), Positives = 21/41 (51%), Gaps = 2/41 (4%)
Query 88 EKGNIYNQ--AAQVWNHTFYFNCLGPPSEETAAPLGPTLKR 126
E+G + N+ ++W+HTFY P T APL P R
Sbjct 26 EQGMVTNRDDMEKIWHHTFYNKLQEHPVLLTEAPLNPKTSR 66
> CE17285
Length=531
Score = 28.1 bits (61), Expect = 8.3, Method: Composition-based stats.
Identities = 17/52 (32%), Positives = 26/52 (50%), Gaps = 4/52 (7%)
Query 78 HQKLETIIKNEKGNIYNQAAQVWNHTFYFNCLGPP--SEETAAPLGPTLKRL 127
HQ ++KN+ G+ + A +WN T N PP ++ P PTL+ L
Sbjct 86 HQLCGLLMKNDCGDFVDPLATIWNMTIPGN--QPPFVPKQVVPPGNPTLRAL 135
> Hs17442535
Length=273
Score = 28.1 bits (61), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 15/39 (38%), Positives = 21/39 (53%), Gaps = 4/39 (10%)
Query 88 EKGNIYNQAAQVWNHTFYFN-CLGP---PSEETAAPLGP 122
E G ++ ++W+HTFY C+ P P T APL P
Sbjct 66 EHGTNWDDMEKIWHHTFYNKLCMTPEEHPMLLTQAPLNP 104
Lambda K H
0.317 0.132 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2707167450
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40