bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0441_orf2
Length=123
Score E
Sequences producing significant alignments: (Bits) Value
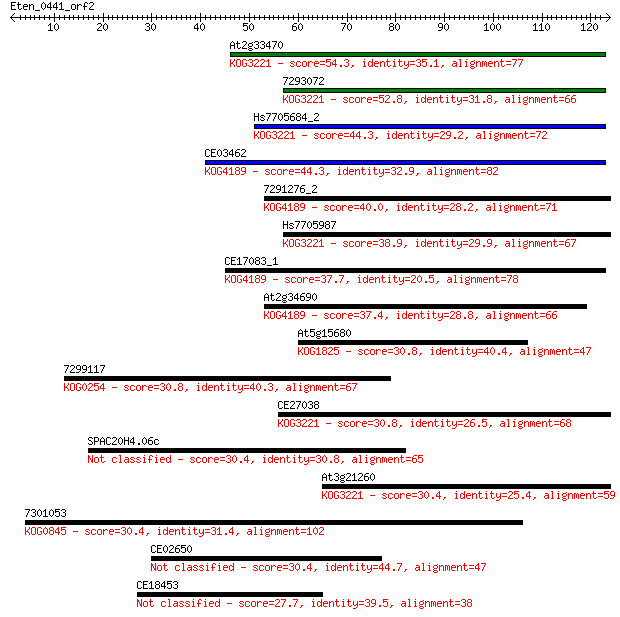
At2g33470 54.3 6e-08
7293072 52.8 2e-07
Hs7705684_2 44.3 5e-05
CE03462 44.3 6e-05
7291276_2 40.0 0.001
Hs7705987 38.9 0.002
CE17083_1 37.7 0.005
At2g34690 37.4 0.006
At5g15680 30.8 0.58
7299117 30.8 0.58
CE27038 30.8 0.72
SPAC20H4.06c 30.4 0.80
At3g21260 30.4 0.81
7301053 30.4 0.85
CE02650 30.4 0.87
CE18453 27.7 5.3
> At2g33470
Length=202
Score = 54.3 bits (129), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 27/77 (35%), Positives = 44/77 (57%), Gaps = 1/77 (1%)
Query 46 KMRKDQNSGVKGMLWMKRALDFVFTLICNTFGTMKDATVKECALDAYDRVLKPYHSFLVS 105
K+ K +S G+LW+ RA+DF+ L N + + CA D+Y + LK +H +L S
Sbjct 85 KIAKGSSSCTNGLLWLTRAMDFLVELFRNLVAHQDWSMPQACA-DSYQKTLKKWHGWLAS 143
Query 106 NIVSLAFSLAPSKDEFV 122
+ S+A LAP + +F+
Sbjct 144 STFSMALKLAPDRKKFM 160
> 7293072
Length=205
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 21/67 (31%), Positives = 38/67 (56%), Gaps = 1/67 (1%)
Query 57 GMLWMKRALDFVFTLICNTFGTMK-DATVKECALDAYDRVLKPYHSFLVSNIVSLAFSLA 115
+LW+KR L + T N + + +K+ DAY+R LKPYH F+V + + + +S
Sbjct 97 ALLWLKRGLQLICTFFENIYNDAQAKEALKQHLQDAYERTLKPYHGFIVQSTIKIIYSWV 156
Query 116 PSKDEFV 122
P++ + +
Sbjct 157 PTRSQLL 163
> Hs7705684_2
Length=181
Score = 44.3 bits (103), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 21/72 (29%), Positives = 39/72 (54%), Gaps = 2/72 (2%)
Query 51 QNSGVKGMLWMKRALDFVFTLICNTFGTMKDATVKECALDAYDRVLKPYHSFLVSNIVSL 110
+NS + +LW+KR L F+ + KD ++ +AY + L+ +H ++V + +L
Sbjct 72 RNSATEALLWLKRGLKFLKGFLTEVKNGEKD--IQTALNNAYGKTLRQHHGWVVRGVFAL 129
Query 111 AFSLAPSKDEFV 122
A PS ++FV
Sbjct 130 ALRATPSYEDFV 141
> CE03462
Length=213
Score = 44.3 bits (103), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 27/82 (32%), Positives = 40/82 (48%), Gaps = 1/82 (1%)
Query 41 ELGIPKMRKDQNSGVKGMLWMKRALDFVFTLICNTFGTMKDATVKECALDAYDRVLKPYH 100
E G + +K SG ++ + RAL+FV L+ F T D V A +YD+ L H
Sbjct 87 ENGTIRNQKPNRSGTGHLMVLNRALEFVIDLLDGVF-TSNDENVSTIARSSYDKHLSQLH 145
Query 101 SFLVSNIVSLAFSLAPSKDEFV 122
S+ + V+ A P K EF+
Sbjct 146 SWPIRTAVAAALYTLPRKPEFL 167
> 7291276_2
Length=204
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 20/71 (28%), Positives = 34/71 (47%), Gaps = 0/71 (0%)
Query 53 SGVKGMLWMKRALDFVFTLICNTFGTMKDATVKECALDAYDRVLKPYHSFLVSNIVSLAF 112
SG + +L + R LDFV+ + D + +AYD L +HSFL+ LA
Sbjct 94 SGSRTLLRLHRGLDFVYEFLNRIQAIPDDQKTVDVCKEAYDDTLGKHHSFLIRKGARLAM 153
Query 113 SLAPSKDEFVR 123
P++ + ++
Sbjct 154 YAMPTRGDLLK 164
> Hs7705987
Length=209
Score = 38.9 bits (89), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 20/71 (28%), Positives = 35/71 (49%), Gaps = 4/71 (5%)
Query 57 GMLWMKRALDFVFTLICNTFGTMKDAT----VKECALDAYDRVLKPYHSFLVSNIVSLAF 112
++W+KR L F+ + + +D ++ A AY+ LK YH ++V I A
Sbjct 93 ALMWLKRGLRFIQVFLQSICDGERDENHPNLIRVNATKAYEMALKKYHGWIVQKIFQAAL 152
Query 113 SLAPSKDEFVR 123
AP K +F++
Sbjct 153 YAAPYKSDFLK 163
> CE17083_1
Length=450
Score = 37.7 bits (86), Expect = 0.005, Method: Composition-based stats.
Identities = 16/78 (20%), Positives = 37/78 (47%), Gaps = 0/78 (0%)
Query 45 PKMRKDQNSGVKGMLWMKRALDFVFTLICNTFGTMKDATVKECALDAYDRVLKPYHSFLV 104
P + + SG +L + RAL+F+ + D ++ + + YD L +H +++
Sbjct 159 PMEKNGKESGAVAILHLNRALEFIVEFMYAAVAATNDDSIPKICKECYDGTLAKHHPWII 218
Query 105 SNIVSLAFSLAPSKDEFV 122
V +A P++++ +
Sbjct 219 RTAVKVAVYTLPTREKML 236
> At2g34690
Length=206
Score = 37.4 bits (85), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 19/66 (28%), Positives = 33/66 (50%), Gaps = 0/66 (0%)
Query 53 SGVKGMLWMKRALDFVFTLICNTFGTMKDATVKECALDAYDRVLKPYHSFLVSNIVSLAF 112
S + +L +KR LD V L + D ++K+ A +Y +V P+H + + VSL
Sbjct 96 SHTRNLLRVKRGLDMVKVLFEQIIASEGDNSLKDPATKSYAQVFAPHHGWAIRKAVSLGM 155
Query 113 SLAPSK 118
P++
Sbjct 156 YALPTR 161
> At5g15680
Length=2163
Score = 30.8 bits (68), Expect = 0.58, Method: Compositional matrix adjust.
Identities = 19/49 (38%), Positives = 26/49 (53%), Gaps = 2/49 (4%)
Query 60 WMKR-ALDFVFTLICNTFGT-MKDATVKECALDAYDRVLKPYHSFLVSN 106
WM R A+ F+F +I F M+D + ALD RVL+ Y S S+
Sbjct 313 WMNRLAIRFLFLVIIENFAPYMQDKNHRYMALDCLHRVLRFYLSVYASS 361
> 7299117
Length=559
Score = 30.8 bits (68), Expect = 0.58, Method: Composition-based stats.
Identities = 27/79 (34%), Positives = 38/79 (48%), Gaps = 12/79 (15%)
Query 12 RMREAAKALRNSRG---GSVVTA------DELI---AWELKELGIPKMRKDQNSGVKGML 59
R REA K+L+ RG ++ A DELI A EL GIP + + G + +
Sbjct 86 RFREAVKSLQWLRGWVPEHMIEAEFNQLYDELITQKAIELSADGIPPPGQRRTLGQRLRM 145
Query 60 WMKRALDFVFTLICNTFGT 78
W KR+ F L+ +F T
Sbjct 146 WRKRSFLVPFLLVSFSFFT 164
> CE27038
Length=231
Score = 30.8 bits (68), Expect = 0.72, Method: Compositional matrix adjust.
Identities = 18/75 (24%), Positives = 33/75 (44%), Gaps = 7/75 (9%)
Query 56 KGMLWMKRALDFVFTLICNTF-----GTMKDAT--VKECALDAYDRVLKPYHSFLVSNIV 108
+G+LW+KR L F+ L+ G +D T + AY + LK +H F+
Sbjct 112 EGLLWLKRGLQFMLELLTEMVTAYNSGLPRDKTEDLSGAVATAYGKSLKRHHGFIAKQAF 171
Query 109 SLAFSLAPSKDEFVR 123
+ P + + ++
Sbjct 172 KVVTMAVPYRRQILK 186
> SPAC20H4.06c
Length=534
Score = 30.4 bits (67), Expect = 0.80, Method: Composition-based stats.
Identities = 20/65 (30%), Positives = 33/65 (50%), Gaps = 6/65 (9%)
Query 17 AKALRNSRGGSVVTADELIAWELKELGIPKMRKDQNSGVKGMLWMKRALDFVFTLICNTF 76
AK LR G V+ D + ++ +LG PKMR D++ + + + FT + +TF
Sbjct 195 AKNLRGGFGSGVLNDDGIEDQDIYDLGAPKMRYDKSINI------TKKVASPFTPVKHTF 248
Query 77 GTMKD 81
+ KD
Sbjct 249 LSKKD 253
> At3g21260
Length=103
Score = 30.4 bits (67), Expect = 0.81, Method: Compositional matrix adjust.
Identities = 15/62 (24%), Positives = 29/62 (46%), Gaps = 7/62 (11%)
Query 65 LDFVFTLICNTFGTMK---DATVKECALDAYDRVLKPYHSFLVSNIVSLAFSLAPSKDEF 121
+DF L+ M + ++EC Y+ +KP+H ++ S +A L P+ + F
Sbjct 1 MDFTLALLQRLVKDMSQNMEQAIEEC----YNLTIKPWHGWISSAAFKVALKLVPNNNTF 56
Query 122 VR 123
+
Sbjct 57 IN 58
> 7301053
Length=319
Score = 30.4 bits (67), Expect = 0.85, Method: Compositional matrix adjust.
Identities = 32/106 (30%), Positives = 43/106 (40%), Gaps = 6/106 (5%)
Query 4 LYIASSSGRMREAAKALRNSRGGSVVTADELIAWE---LKELGIPKMRKDQNSGVKGMLW 60
L+I + R R + L+ R SV L A E ++ELGIP D VK
Sbjct 68 LHIKQQTQRERAVQQMLQ--RNVSVSAKVALYAEERFIVEELGIPMSWVDYAKAVKAGAS 125
Query 61 MKRALDFVFTLICNTFGTMKDATVKECALDA-YDRVLKPYHSFLVS 105
K L + L F T D + A DA + +K HS L+
Sbjct 126 GKHHLQAKYLLKAKHFATAHDVIFQHIAPDAIINGKMKYLHSLLIQ 171
> CE02650
Length=360
Score = 30.4 bits (67), Expect = 0.87, Method: Compositional matrix adjust.
Identities = 21/47 (44%), Positives = 23/47 (48%), Gaps = 8/47 (17%)
Query 30 TADELIAWELKELGIPKMRKDQNSGVKGMLWMKRALDFVFTLICNTF 76
TA+EL W GI K QNS +WMK FVF LI TF
Sbjct 8 TAEELETWRCTSDGI---FKSQNS-----IWMKINFVFVFILIFLTF 46
> CE18453
Length=244
Score = 27.7 bits (60), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 15/47 (31%), Positives = 24/47 (51%), Gaps = 9/47 (19%)
Query 27 SVVTADELIA-------WELKELG--IPKMRKDQNSGVKGMLWMKRA 64
S TAD+++ WE + L +PK ++ N+G+ LW K A
Sbjct 188 SKATADDIVGRILTGGQWEREHLPPVVPKAQRPMNNGINRHLWKKEA 234
Lambda K H
0.321 0.133 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1191192512
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40