bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0460_orf1
Length=402
Score E
Sequences producing significant alignments: (Bits) Value
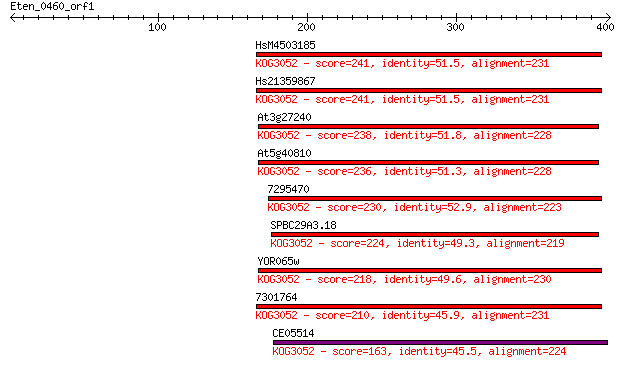
HsM4503185 241 2e-63
Hs21359867 241 2e-63
At3g27240 238 1e-62
At5g40810 236 5e-62
7295470 230 5e-60
SPBC29A3.18 224 2e-58
YOR065w 218 2e-56
7301764 210 4e-54
CE05514 163 7e-40
> HsM4503185
Length=325
Score = 241 bits (614), Expect = 2e-63, Method: Compositional matrix adjust.
Identities = 119/231 (51%), Positives = 159/231 (68%), Gaps = 0/231 (0%)
Query 166 PPHYPFWFKSIFHSHDIPSVRRGYEVYRKVCSTCHSMEQLHFRHLVGEVLPELRVKQIAA 225
PP YP+ + + S D S+RRG++VY++VC++CHSM+ + +RHLVG E K++AA
Sbjct 91 PPSYPWSHRGLLSSLDHTSIRRGFQVYKQVCASCHSMDFVAYRHLVGVCYTEDEAKELAA 150
Query 226 EYDVEDGPNDSGEMYTRPGVAGDAFPKPYPNEEAARFANNGAYPPDLSLITGARMHGPDY 285
E +V+DGPN+ GEM+ RPG D FPKPYPN EAAR ANNGA PPDLS I AR G DY
Sbjct 151 EVEVQDGPNEDGEMFMRPGKLFDYFPKPYPNSEAARAANNGALPPDLSYIVRARHGGEDY 210
Query 286 IMALLNGYHEPPMGVELRPGLYWNVWFPGNAIAMAPPLQDEMIEYEDGTPCNVSQMSKDV 345
+ +LL GY EPP GV LR GLY+N +FPG AIAMAPP+ +++E++DGTP +SQ++KDV
Sbjct 211 VFSLLTGYCEPPTGVSLREGLYFNPYFPGQAIAMAPPIYTDVLEFDDGTPATMSQIAKDV 270
Query 346 VNFLTWATEPTCDERKLFGLKAVSAALLGCALMTVWWRFFWVIYATRRIDF 396
FL WA+EP D RK GLK + L L+ R W + +R++ +
Sbjct 271 CTFLRWASEPEHDHRKRMGLKMLMMMALLVPLVYTIKRHKWSVLKSRKLAY 321
> Hs21359867
Length=325
Score = 241 bits (614), Expect = 2e-63, Method: Compositional matrix adjust.
Identities = 119/231 (51%), Positives = 159/231 (68%), Gaps = 0/231 (0%)
Query 166 PPHYPFWFKSIFHSHDIPSVRRGYEVYRKVCSTCHSMEQLHFRHLVGEVLPELRVKQIAA 225
PP YP+ + + S D S+RRG++VY++VC++CHSM+ + +RHLVG E K++AA
Sbjct 91 PPSYPWSHRGLLSSLDHTSIRRGFQVYKQVCASCHSMDFVAYRHLVGVCYTEDEAKELAA 150
Query 226 EYDVEDGPNDSGEMYTRPGVAGDAFPKPYPNEEAARFANNGAYPPDLSLITGARMHGPDY 285
E +V+DGPN+ GEM+ RPG D FPKPYPN EAAR ANNGA PPDLS I AR G DY
Sbjct 151 EVEVQDGPNEDGEMFMRPGKLFDYFPKPYPNSEAARAANNGALPPDLSYIVRARHGGEDY 210
Query 286 IMALLNGYHEPPMGVELRPGLYWNVWFPGNAIAMAPPLQDEMIEYEDGTPCNVSQMSKDV 345
+ +LL GY EPP GV LR GLY+N +FPG AIAMAPP+ +++E++DGTP +SQ++KDV
Sbjct 211 VFSLLTGYCEPPTGVSLREGLYFNPYFPGQAIAMAPPIYTDVLEFDDGTPATMSQIAKDV 270
Query 346 VNFLTWATEPTCDERKLFGLKAVSAALLGCALMTVWWRFFWVIYATRRIDF 396
FL WA+EP D RK GLK + L L+ R W + +R++ +
Sbjct 271 CTFLRWASEPEHDHRKRMGLKMLMMMALLVPLVYTIKRHKWSVLKSRKLAY 321
> At3g27240
Length=307
Score = 238 bits (608), Expect = 1e-62, Method: Compositional matrix adjust.
Identities = 118/228 (51%), Positives = 153/228 (67%), Gaps = 0/228 (0%)
Query 167 PHYPFWFKSIFHSHDIPSVRRGYEVYRKVCSTCHSMEQLHFRHLVGEVLPELRVKQIAAE 226
P YP+ I S+D S+RRG++VY++VC++CHSM + +R LVG E K +AAE
Sbjct 74 PEYPWPHDGILSSYDHASIRRGHQVYQQVCASCHSMSLISYRDLVGVAYTEEEAKAMAAE 133
Query 227 YDVEDGPNDSGEMYTRPGVAGDAFPKPYPNEEAARFANNGAYPPDLSLITGARMHGPDYI 286
+V DGPND GEM+TRPG D FP+PY NE AARFAN GAYPPDLSLIT AR +GP+Y+
Sbjct 134 IEVVDGPNDEGEMFTRPGKLSDRFPQPYANESAARFANGGAYPPDLSLITKARHNGPNYV 193
Query 287 MALLNGYHEPPMGVELRPGLYWNVWFPGNAIAMAPPLQDEMIEYEDGTPCNVSQMSKDVV 346
ALL GY +PP G+ +R GL++N +FPG AIAM L DE +EYEDG P +QM KD+V
Sbjct 194 FALLTGYRDPPAGISIREGLHYNPYFPGGAIAMPKMLNDEAVEYEDGVPATEAQMGKDIV 253
Query 347 NFLTWATEPTCDERKLFGLKAVSAALLGCALMTVWWRFFWVIYATRRI 394
+FL WA EP +ERKL G K + L + R W + +R++
Sbjct 254 SFLAWAAEPEMEERKLMGFKWIFLLSLALLQAAYYRRLKWSVLKSRKL 301
> At5g40810
Length=307
Score = 236 bits (603), Expect = 5e-62, Method: Compositional matrix adjust.
Identities = 117/228 (51%), Positives = 155/228 (67%), Gaps = 0/228 (0%)
Query 167 PHYPFWFKSIFHSHDIPSVRRGYEVYRKVCSTCHSMEQLHFRHLVGEVLPELRVKQIAAE 226
P+YP+ + I S+D S+RRG++VY++VC++CHSM + +R LVG E K +AAE
Sbjct 74 PNYPWPHEGILSSYDHASIRRGHQVYQQVCASCHSMSLISYRDLVGVAYTEEEAKAMAAE 133
Query 227 YDVEDGPNDSGEMYTRPGVAGDAFPKPYPNEEAARFANNGAYPPDLSLITGARMHGPDYI 286
+V DGPND GEM+TRPG D P+PY NE AARFAN GAYPPDLSL+T AR +G +Y+
Sbjct 134 IEVVDGPNDEGEMFTRPGKLSDRLPEPYSNESAARFANGGAYPPDLSLVTKARHNGQNYV 193
Query 287 MALLNGYHEPPMGVELRPGLYWNVWFPGNAIAMAPPLQDEMIEYEDGTPCNVSQMSKDVV 346
ALL GY +PP G+ +R GL++N +FPG AIAM L DE +EYEDGTP +QM KDVV
Sbjct 194 FALLTGYRDPPAGISIREGLHYNPYFPGGAIAMPKMLNDEAVEYEDGTPATEAQMGKDVV 253
Query 347 NFLTWATEPTCDERKLFGLKAVSAALLGCALMTVWWRFFWVIYATRRI 394
+FL+WA EP +ERKL G K + L + R W + +R++
Sbjct 254 SFLSWAAEPEMEERKLMGFKWIFLLSLALLQAAYYRRLKWSVLKSRKL 301
> 7295470
Length=307
Score = 230 bits (586), Expect = 5e-60, Method: Compositional matrix adjust.
Identities = 118/225 (52%), Positives = 149/225 (66%), Gaps = 4/225 (1%)
Query 174 KSIFHSHDIPSVRRGYEVYRKVCSTCHSMEQLHFRHLVGEVLPELRVKQIAAEYDVEDGP 233
K +F + D SVRRGYEVY++VCS CHSM+ + +R+LVG E K A + V+DGP
Sbjct 80 KGLFDALDHQSVRRGYEVYKQVCSACHSMQYIAYRNLVGVTHTEAEAKAEAEQITVKDGP 139
Query 234 NDSGEMYTRPGVAGDAFPKPYPNEEAARFANNGAYPPDLSLITGARMHGPDYIMALLNGY 293
+D+G YTRPG D FP PYPNEEAAR ANNGAYPPDLS I AR G DYI +LL GY
Sbjct 140 DDTGNYYTRPGKLSDYFPSPYPNEEAARAANNGAYPPDLSYIVSARKGGEDYIFSLLTGY 199
Query 294 HEPPMGVELRPGLYWNVWFPGNAIAMAPPLQDEMIEYEDGTPCNVSQMSKDVVNFLTWAT 353
H+ P GV LR G Y+N +FPG AI+MA L +E+IEYEDGTP SQ++KDV FL W +
Sbjct 200 HDAPAGVVLREGQYFNPYFPGGAISMAQVLYNEVIEYEDGTPPTQSQLAKDVATFLKWTS 259
Query 354 EPTCDERKLFGLKAVSAALLGCALMTVWW--RFFWVIYATRRIDF 396
EP D+RK +K + +LG + ++ R W +R+I F
Sbjct 260 EPEHDDRKQLLIKVI--GILGFLTVISYYIKRHKWSSLKSRKIVF 302
> SPBC29A3.18
Length=307
Score = 224 bits (572), Expect = 2e-58, Method: Compositional matrix adjust.
Identities = 108/219 (49%), Positives = 140/219 (63%), Gaps = 0/219 (0%)
Query 176 IFHSHDIPSVRRGYEVYRKVCSTCHSMEQLHFRHLVGEVLPELRVKQIAAEYDVEDGPND 235
+ D S+RRG++VYR+VCS CHS+ + +RHLVG KQ+A+E + EDGP+D
Sbjct 82 VLSGFDHASLRRGFQVYREVCSACHSLNLIAWRHLVGVTHTADEAKQMASEVEYEDGPDD 141
Query 236 SGEMYTRPGVAGDAFPKPYPNEEAARFANNGAYPPDLSLITGARMHGPDYIMALLNGYHE 295
G M+ RPG D P PYPN EAAR +NNGA PPDLS + R G DYI +LL GY E
Sbjct 142 EGNMFKRPGKLSDFLPPPYPNVEAARASNNGAAPPDLSCVVRGRHGGQDYIYSLLTGYTE 201
Query 296 PPMGVELRPGLYWNVWFPGNAIAMAPPLQDEMIEYEDGTPCNVSQMSKDVVNFLTWATEP 355
PP GVE+ G+ +N +FPG IAMA PL D+ +E+EDGTP +Q +KDVVNFL WA+EP
Sbjct 202 PPAGVEVPDGMNFNPFFPGTQIAMARPLFDDAVEFEDGTPATTAQAAKDVVNFLHWASEP 261
Query 356 TCDERKLFGLKAVSAALLGCALMTVWWRFFWVIYATRRI 394
D RK G + ++ + AL + RF W R+I
Sbjct 262 ELDIRKKMGFQVITVLTILTALSMWYKRFKWTPIKNRKI 300
> YOR065w
Length=309
Score = 218 bits (554), Expect = 2e-56, Method: Compositional matrix adjust.
Identities = 114/232 (49%), Positives = 145/232 (62%), Gaps = 3/232 (1%)
Query 167 PHYPFWFKSIFHSHDIPSVRRGYEVYRKVCSTCHSMEQLHFRHLVGEVLPELRVKQIAAE 226
P Y + F + D S+RRGY+VYR+VC+ CHS++++ +R LVG V+ +A E
Sbjct 72 PAYAWSHNGPFETFDHASIRRGYQVYREVCAACHSLDRVAWRTLVGVSHTNEEVRNMAEE 131
Query 227 YDVEDGPNDSGEMYTRPGVAGDAFPKPYPNEEAARFANNGAYPPDLSLITGARMHGPDYI 286
++ +D P++ G RPG D P PYPNE+AAR AN GA PPDLSLI AR G DYI
Sbjct 132 FEYDDEPDEQGNPKKRPGKLSDYIPGPYPNEQAARAANQGALPPDLSLIVKARHGGCDYI 191
Query 287 MALLNGY-HEPPMGVELRPGLYWNVWFPGNAIAMAPPLQDEMIEYEDGTPCNVSQMSKDV 345
+LL GY EPP GV L PG +N +FPG +IAMA L D+M+EYEDGTP SQM+KDV
Sbjct 192 FSLLTGYPDEPPAGVALPPGSNYNPYFPGGSIAMARVLFDDMVEYEDGTPATTSQMAKDV 251
Query 346 VNFLTWATEPTCDERKLFGLKAVSAALLGCALMTVWW-RFFWVIYATRRIDF 396
FL W EP DERK GLK V L L+++W +F W TR+ F
Sbjct 252 TTFLNWCAEPEHDERKRLGLKTV-IILSSLYLLSIWVKKFKWAGIKTRKFVF 302
> 7301764
Length=344
Score = 210 bits (535), Expect = 4e-54, Method: Compositional matrix adjust.
Identities = 106/232 (45%), Positives = 141/232 (60%), Gaps = 1/232 (0%)
Query 166 PPHYPFWFKSIFHSHDIPSVRRGYEVYRKVCSTCHSMEQLHFRHLVGEVLPELRVKQIAA 225
P H + K + + D SVRRGY VY++VCS+CHS++ + +R+LVG + E K A
Sbjct 75 PAHQHWNHKGLLSALDKESVRRGYTVYKEVCSSCHSLQYMAYRNLVGVCMTEAEAKAEAE 134
Query 226 EYDVEDGPNDSGEMYTRPGVAGDAFPKPYPNEEAARFANNGAYPPDLSLITGARMHGPDY 285
V DGPN+ GE Y RPG D FP PY NEEAAR ANNG+YPPDLS I AR G DY
Sbjct 135 AITVRDGPNEEGEYYERPGKLSDHFPSPYANEEAARSANNGSYPPDLSYIVSARKGGEDY 194
Query 286 IMALLNGYHEPPMGVELRPGLYWNVWFPGNAIAMAPPLQDEMIEYED-GTPCNVSQMSKD 344
+ +LL GY +PP G LR GLY+N +F G AIAM + +E++ +ED P + +Q++KD
Sbjct 195 VFSLLTGYCDPPAGFALRDGLYFNPYFSGGAIAMGKVVDNEVVSFEDPNVPASAAQIAKD 254
Query 345 VVNFLTWATEPTCDERKLFGLKAVSAALLGCALMTVWWRFFWVIYATRRIDF 396
V FL W +EP DER+L +K + + RF W +R+I F
Sbjct 255 VCVFLKWTSEPETDERRLLLIKVTLISTFLIGISYYIKRFKWSTLKSRKIFF 306
> CE05514
Length=285
Score = 163 bits (412), Expect = 7e-40, Method: Compositional matrix adjust.
Identities = 102/224 (45%), Positives = 135/224 (60%), Gaps = 1/224 (0%)
Query 177 FHSHDIPSVRRGYEVYRKVCSTCHSMEQLHFRHLVGEVLPELRVKQIAAEYDVEDGPNDS 236
F S DI SVRRGYEVY++VC+ CHSM+ LH+RH V ++ E K AA+ + D +D
Sbjct 55 FSSFDIASVRRGYEVYKQVCAACHSMKFLHYRHFVDTIMTEEEAKAEAADALIND-VDDK 113
Query 237 GEMYTRPGVAGDAFPKPYPNEEAARFANNGAYPPDLSLITGARMHGPDYIMALLNGYHEP 296
G RPG+ D P PYPN++AA ANNGA PPDLSL+ AR G DY+ +LL GY E
Sbjct 114 GASIQRPGMLTDKLPNPYPNKKAAAAANNGAAPPDLSLMALARHGGDDYVFSLLTGYLEA 173
Query 297 PMGVELRPGLYWNVWFPGNAIAMAPPLQDEMIEYEDGTPCNVSQMSKDVVNFLTWATEPT 356
P GV++ G +N +FPG I+M L DE IEY+DGTP +SQ +KDV F+ WA EP
Sbjct 174 PAGVKVDDGKAYNPYFPGGIISMPQQLFDEGIEYKDGTPATMSQQAKDVSAFMHWAAEPF 233
Query 357 CDERKLFGLKAVSAALLGCALMTVWWRFFWVIYATRRIDFGKLK 400
D RK + LK + ++ R W +++ F +K
Sbjct 234 HDTRKKWALKIAALIPFVAVVLIYGKRHIWSFTKSQKFLFKTVK 277
Lambda K H
0.322 0.139 0.458
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 10175212506
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40