bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0465_orf1
Length=204
Score E
Sequences producing significant alignments: (Bits) Value
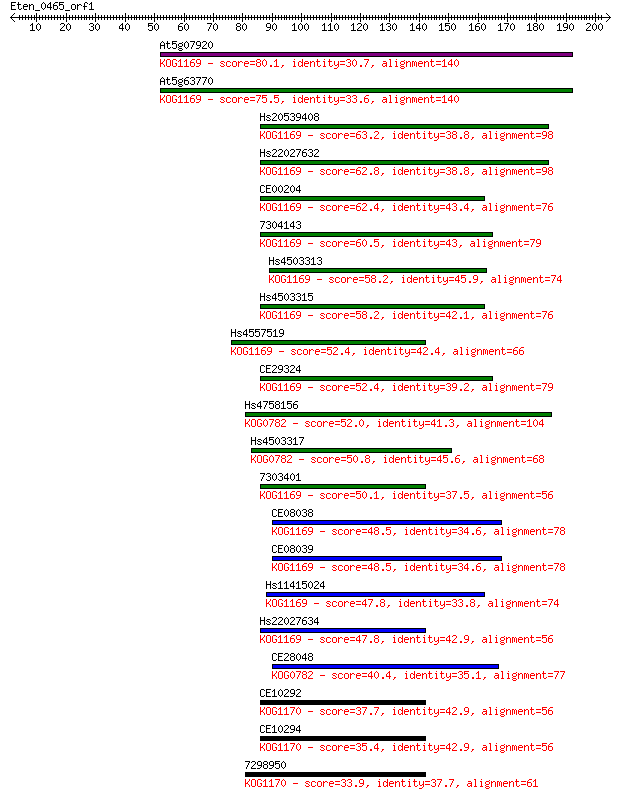
At5g07920 80.1 2e-15
At5g63770 75.5 8e-14
Hs20539408 63.2 4e-10
Hs22027632 62.8 4e-10
CE00204 62.4 6e-10
7304143 60.5 2e-09
Hs4503313 58.2 1e-08
Hs4503315 58.2 1e-08
Hs4557519 52.4 6e-07
CE29324 52.4 6e-07
Hs4758156 52.0 9e-07
Hs4503317 50.8 2e-06
7303401 50.1 3e-06
CE08038 48.5 9e-06
CE08039 48.5 9e-06
Hs11415024 47.8 1e-05
Hs22027634 47.8 2e-05
CE28048 40.4 0.002
CE10292 37.7 0.018
CE10294 35.4 0.089
7298950 33.9 0.22
> At5g07920
Length=728
Score = 80.1 bits (196), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 43/140 (30%), Positives = 81/140 (57%), Gaps = 2/140 (1%)
Query 52 RDQQYMSAPHLGALTASVHRRHQLQQQRFDHWKPQKIDDGLIEVVAFKSLFHLGQVQVGL 111
D + + ++G+ V Q + + ++++ PQ + D ++EVV+ +HLG++QVGL
Sbjct 578 EDAEGILVANIGSYMGGVDL-WQNEDETYENFDPQSMHDKIVEVVSISGTWHLGKLQVGL 636
Query 112 AKALRLCQGRSVCLHVSGSLPLQVDGEPQLLEGSCRIAITHNCQAPVLVASEKSPRMQTL 171
++A RL QG +V + + LP+Q+DGEP + C + I+H+ QA +L + + P
Sbjct 637 SRARRLAQGSAVKIQLCAPLPVQIDGEP-WNQQPCTLTISHHGQAFMLKRAAEEPLGHAA 695
Query 172 SVVQEVLNRARDRRVISKMQ 191
+++ +VL A +VI+ Q
Sbjct 696 AIITDVLENAETNQVINASQ 715
> At5g63770
Length=712
Score = 75.5 bits (184), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 47/143 (32%), Positives = 80/143 (55%), Gaps = 8/143 (5%)
Query 52 RDQQYMSAPHLGALTASVHRRHQLQQQRFDH---WKPQKIDDGLIEVVAFKSLFHLGQVQ 108
+D + + ++G+ V L Q ++H + Q + D +EVV + +HLG++Q
Sbjct 562 KDSEGLIVLNIGSYMGGV----DLWQNDYEHDDNFSIQCMHDKTLEVVCVRGAWHLGKLQ 617
Query 109 VGLAKALRLCQGRSVCLHVSGSLPLQVDGEPQLLEGSCRIAITHNCQAPVLVASEKSPRM 168
VGL++A RL QG+ + +HVS P+Q+DGEP + + C + ITH+ Q +L + PR
Sbjct 618 VGLSQARRLAQGKVIRIHVSSPFPVQIDGEPFIQQPGC-LEITHHGQVFMLRRASDEPRG 676
Query 169 QTLSVVQEVLNRARDRRVISKMQ 191
+++ EVL A + VI+ Q
Sbjct 677 HAAAIMNEVLLDAECKGVINASQ 699
> Hs20539408
Length=738
Score = 63.2 bits (152), Expect = 4e-10, Method: Composition-based stats.
Identities = 38/100 (38%), Positives = 56/100 (56%), Gaps = 5/100 (5%)
Query 86 QKIDDGLIEVVAFKSLFHLGQVQVGLAKA-LRLCQGRSVCLHVSGSLPLQVDGEPQLLEG 144
Q + D L+EVV + +GQ+ GL A RL Q V + S SLP+Q+DGEP ++
Sbjct 642 QDLSDQLLEVVGLEGAMEMGQIYTGLKSAGRRLAQCSCVVIRTSKSLPMQIDGEP-WMQT 700
Query 145 SCRIAITHNCQAPVLVASEKSPRMQTL-SVVQEVLNRARD 183
C I ITH QAP+L+ P+ S+V+ NR+++
Sbjct 701 PCTIKITHKNQAPMLMGP--PPKTGLFCSLVKRTRNRSKE 738
> Hs22027632
Length=804
Score = 62.8 bits (151), Expect = 4e-10, Method: Composition-based stats.
Identities = 38/100 (38%), Positives = 56/100 (56%), Gaps = 5/100 (5%)
Query 86 QKIDDGLIEVVAFKSLFHLGQVQVGLAKA-LRLCQGRSVCLHVSGSLPLQVDGEPQLLEG 144
Q + D L+EVV + +GQ+ GL A RL Q V + S SLP+Q+DGEP ++
Sbjct 708 QDLSDQLLEVVGLEGAMEMGQIYTGLKSAGRRLAQCSCVVIRTSKSLPMQIDGEP-WMQT 766
Query 145 SCRIAITHNCQAPVLVASEKSPRMQTL-SVVQEVLNRARD 183
C I ITH QAP+L+ P+ S+V+ NR+++
Sbjct 767 PCTIKITHKNQAPMLMGP--PPKTGLFCSLVKRTRNRSKE 804
> CE00204
Length=827
Score = 62.4 bits (150), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 33/76 (43%), Positives = 46/76 (60%), Gaps = 1/76 (1%)
Query 86 QKIDDGLIEVVAFKSLFHLGQVQVGLAKALRLCQGRSVCLHVSGSLPLQVDGEPQLLEGS 145
Q I DGLIE+V +S +GQ++ G+ A RL Q +V + S P+Q+DGEP ++
Sbjct 715 QDIGDGLIELVGLESAMQMGQIKAGVRGARRLSQCSTVVIQTHKSFPMQIDGEP-WMQPP 773
Query 146 CRIAITHNCQAPVLVA 161
C I ITH QA +LV
Sbjct 774 CIIQITHKNQAKMLVG 789
> 7304143
Length=791
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 34/82 (41%), Positives = 48/82 (58%), Gaps = 4/82 (4%)
Query 86 QKIDDGLIEVVAFKSLFHLGQVQVGL-AKALRLCQGRSVCLHVSGSLPLQVDGEPQLLEG 144
Q D LIEV+ ++ H+GQV+ GL A RL Q V + + P+Q+DGEP ++
Sbjct 701 QDFGDRLIEVIGLENCLHMGQVRTGLRASGRRLAQCSEVIIKTKKTFPMQIDGEP-WMQM 759
Query 145 SCRIAITHNCQAPVLVA--SEK 164
C I +TH Q P+L+A SEK
Sbjct 760 PCTIKVTHKNQVPMLMAPRSEK 781
> Hs4503313
Length=567
Score = 58.2 bits (139), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 34/75 (45%), Positives = 44/75 (58%), Gaps = 2/75 (2%)
Query 89 DDGLIEVVAFKSLFHLGQVQVGLAKALRLCQGRSVCLHVSGS-LPLQVDGEPQLLEGSCR 147
DDGL+EVV FH Q+QV LA R+ Q +V L + S +P+QVDGEP +G C
Sbjct 473 DDGLLEVVGVYGSFHCAQIQVKLANPFRIGQAHTVRLILKCSMMPMQVDGEPW-AQGPCT 531
Query 148 IAITHNCQAPVLVAS 162
+ ITH A +L S
Sbjct 532 VTITHKTHAMMLYFS 546
> Hs4503315
Length=791
Score = 58.2 bits (139), Expect = 1e-08, Method: Composition-based stats.
Identities = 32/77 (41%), Positives = 45/77 (58%), Gaps = 2/77 (2%)
Query 86 QKIDDGLIEVVAFKSLFHLGQVQVGLAKA-LRLCQGRSVCLHVSGSLPLQVDGEPQLLEG 144
Q + D L+EVV + +GQ+ GL A RL Q SV + + LP+QVDGEP ++
Sbjct 698 QDLSDQLLEVVGLEGAMEMGQIYTGLKSAGRRLAQCASVTIRTNKLLPMQVDGEP-WMQP 756
Query 145 SCRIAITHNCQAPVLVA 161
C I ITH QAP+++
Sbjct 757 CCTIKITHKNQAPMMMG 773
> Hs4557519
Length=942
Score = 52.4 bits (124), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 28/66 (42%), Positives = 41/66 (62%), Gaps = 3/66 (4%)
Query 76 QQQRFDHWKPQKIDDGLIEVVAFKSLFHLGQVQVGLAKALRLCQGRSVCLHVSGSLPLQV 135
RF+ KP ++DDGL+EVV + H+GQVQ GL +R+ QG + + + P+QV
Sbjct 833 SDTRFE--KP-RMDDGLLEVVGVTGVVHMGQVQGGLRSGIRIAQGSYFRVTLLKATPVQV 889
Query 136 DGEPQL 141
DGEP +
Sbjct 890 DGEPWV 895
> CE29324
Length=503
Score = 52.4 bits (124), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 31/81 (38%), Positives = 46/81 (56%), Gaps = 3/81 (3%)
Query 86 QKIDDGLIEVVAFKSLFHLGQVQVGLAKALRLCQGRSVCLHVSG--SLPLQVDGEPQLLE 143
Q+ DD +IEV A S FH+ Q+Q+GLA L + Q + L G S P+Q DGE +
Sbjct 412 QECDDEMIEVFAVTSSFHIAQMQIGLASPLCIGQAKHAKLVFKGNHSFPMQSDGEAW-VN 470
Query 144 GSCRIAITHNCQAPVLVASEK 164
+ + I+H C+ +L +EK
Sbjct 471 SAGTVLISHKCKTAMLRKAEK 491
> Hs4758156
Length=1065
Score = 52.0 bits (123), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 43/111 (38%), Positives = 60/111 (54%), Gaps = 15/111 (13%)
Query 81 DH--WKPQKIDDGLIEVVAFKSLFHLGQVQVGLAKALRLCQGRSVCLHVSGSLPLQVDGE 138
DH ++PQ+ DDG IEV+ F ++ L +QVG RL Q R V L S+P+QVDGE
Sbjct 625 DHHDFEPQRHDDGYIEVIGF-TMASLAALQVG-GHGERLHQCREVMLLTYKSIPMQVDGE 682
Query 139 PQLLEGSCRIA-----ITHNCQAPVLVASEKSPRMQTLSVVQEVLNRARDR 184
P CR+A I+ QA ++ S++ M L+ Q V +R R R
Sbjct 683 P------CRLAPAMIRISLRNQANMVQKSKRRTSMPLLNDPQSVPDRLRIR 727
> Hs4503317
Length=928
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 31/69 (44%), Positives = 43/69 (62%), Gaps = 3/69 (4%)
Query 83 WKPQKIDDGLIEVVAFKSLFHLGQVQVGLAKALRLCQGRSVCLHVSGSLPLQVDGEPQLL 142
++PQ+ DDG +EV+ F ++ L +QVG RL Q R V L S ++P+QVDGEP L
Sbjct 549 FEPQRHDDGYLEVIGF-TMTSLAALQVG-GHGERLTQCREVVLTTSKAIPVQVDGEPCKL 606
Query 143 EGS-CRIAI 150
S RIA+
Sbjct 607 AASRIRIAL 615
> 7303401
Length=534
Score = 50.1 bits (118), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 21/56 (37%), Positives = 35/56 (62%), Gaps = 0/56 (0%)
Query 86 QKIDDGLIEVVAFKSLFHLGQVQVGLAKALRLCQGRSVCLHVSGSLPLQVDGEPQL 141
I DG++EV S FH+ Q+Q ++K +R+ Q + + L V ++P+Q DGEP +
Sbjct 457 NSISDGMMEVFGIVSSFHIAQLQCNISKPVRIGQAKQIRLQVKETVPMQADGEPWM 512
> CE08038
Length=950
Score = 48.5 bits (114), Expect = 9e-06, Method: Composition-based stats.
Identities = 27/78 (34%), Positives = 41/78 (52%), Gaps = 0/78 (0%)
Query 90 DGLIEVVAFKSLFHLGQVQVGLAKALRLCQGRSVCLHVSGSLPLQVDGEPQLLEGSCRIA 149
DGL+EVV + LG +Q LA +R+ QG S+ + P+QVDGEP +
Sbjct 818 DGLLEVVGISDVSRLGLIQSKLAAGIRIAQGGSIRITTHEEWPVQVDGEPHIQPPGTITI 877
Query 150 ITHNCQAPVLVASEKSPR 167
+ +A +L ++KS R
Sbjct 878 LKSALKAQMLKKAKKSRR 895
> CE08039
Length=952
Score = 48.5 bits (114), Expect = 9e-06, Method: Composition-based stats.
Identities = 27/78 (34%), Positives = 41/78 (52%), Gaps = 0/78 (0%)
Query 90 DGLIEVVAFKSLFHLGQVQVGLAKALRLCQGRSVCLHVSGSLPLQVDGEPQLLEGSCRIA 149
DGL+EVV + LG +Q LA +R+ QG S+ + P+QVDGEP +
Sbjct 820 DGLLEVVGISDVSRLGLIQSKLAAGIRIAQGGSIRITTHEEWPVQVDGEPHIQPPGTITI 879
Query 150 ITHNCQAPVLVASEKSPR 167
+ +A +L ++KS R
Sbjct 880 LKSALKAQMLKKAKKSRR 897
> Hs11415024
Length=735
Score = 47.8 bits (112), Expect = 1e-05, Method: Composition-based stats.
Identities = 25/75 (33%), Positives = 40/75 (53%), Gaps = 2/75 (2%)
Query 88 IDDGLIEVVAFKSLFHLGQVQVGLAKA-LRLCQGRSVCLHVSGSLPLQVDGEPQLLEGSC 146
+ D +EVV + +GQ+ L A RL + + H + +LP+Q+D EP ++ C
Sbjct 649 LSDKRLEVVGLEGAIEMGQIYTKLKNAGRRLAKCSEITFHTTKTLPMQIDVEP-WMQTPC 707
Query 147 RIAITHNCQAPVLVA 161
I ITH Q P+L+
Sbjct 708 TIKITHKNQMPMLMG 722
> Hs22027634
Length=773
Score = 47.8 bits (112), Expect = 2e-05, Method: Composition-based stats.
Identities = 24/57 (42%), Positives = 33/57 (57%), Gaps = 1/57 (1%)
Query 86 QKIDDGLIEVVAFKSLFHLGQVQVGLAKA-LRLCQGRSVCLHVSGSLPLQVDGEPQL 141
Q + D L+EVV + +GQ+ GL A RL Q V + S SLP+Q+DGEP +
Sbjct 708 QDLSDQLLEVVGLEGAMEMGQIYTGLKSAGRRLAQCSCVVIRTSKSLPMQIDGEPWM 764
> CE28048
Length=937
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 27/77 (35%), Positives = 44/77 (57%), Gaps = 3/77 (3%)
Query 90 DGLIEVVAFKSLFHLGQVQVGLAKALRLCQGRSVCLHVSGSLPLQVDGEPQLLEGSCRIA 149
DG +EV+ F + L +Q+G K R+ Q V + + ++P+QVDGEP LL S I
Sbjct 678 DGKVEVLGFTTA-TLAALQMG-GKGERIAQCSRVRVITNKAIPMQVDGEPCLLAPSI-IT 734
Query 150 ITHNCQAPVLVASEKSP 166
+ + + P+L +K+P
Sbjct 735 LGFHSKVPMLKREKKTP 751
> CE10292
Length=1372
Score = 37.7 bits (86), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 24/61 (39%), Positives = 34/61 (55%), Gaps = 5/61 (8%)
Query 86 QKIDDGLIEVVAFKSLFHLGQVQVGLAKAL---RLCQGRSVCLHVSGS--LPLQVDGEPQ 140
Q DD ++EVVA + H+ +V A L R+ Q R V + + G +P+QVDGEP
Sbjct 940 QSFDDRILEVVALFGVIHVATSRVPNAVRLQNHRIAQCRHVRIVILGDEPIPVQVDGEPW 999
Query 141 L 141
L
Sbjct 1000 L 1000
> CE10294
Length=1321
Score = 35.4 bits (80), Expect = 0.089, Method: Composition-based stats.
Identities = 24/61 (39%), Positives = 34/61 (55%), Gaps = 5/61 (8%)
Query 86 QKIDDGLIEVVAFKSLFHLGQVQVGLAKAL---RLCQGRSVCLHVSGS--LPLQVDGEPQ 140
Q DD ++EVVA + H+ +V A L R+ Q R V + + G +P+QVDGEP
Sbjct 889 QSFDDRILEVVALFGVIHVATSRVPNAVRLQNHRIAQCRHVRIVILGDEPIPVQVDGEPW 948
Query 141 L 141
L
Sbjct 949 L 949
> 7298950
Length=552
Score = 33.9 bits (76), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 37/68 (54%), Gaps = 12/68 (17%)
Query 81 DHWKPQKIDDGLIEVVAFKSLFHLGQVQVGLAKAL-----RLCQGRSVCLHVSGS--LPL 133
D + P DD ++EVVA G VQ+ ++ + R+ Q +SV +++ G +P+
Sbjct 191 DIFLPPSFDDRVLEVVAV-----FGSVQMAASRLINLQHHRIAQCQSVQINILGDEEIPI 245
Query 134 QVDGEPQL 141
QVDGE L
Sbjct 246 QVDGEAWL 253
Lambda K H
0.321 0.132 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3622842326
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40