bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0489_orf3
Length=110
Score E
Sequences producing significant alignments: (Bits) Value
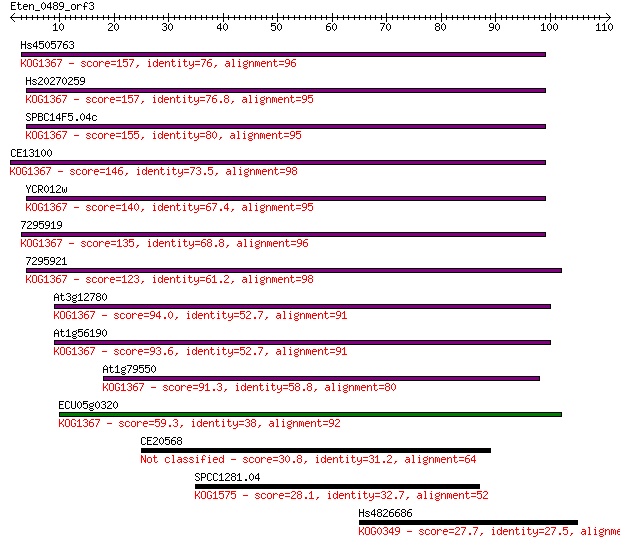
Hs4505763 157 5e-39
Hs20270259 157 6e-39
SPBC14F5.04c 155 1e-38
CE13100 146 8e-36
YCR012w 140 7e-34
7295919 135 3e-32
7295921 123 8e-29
At3g12780 94.0 6e-20
At1g56190 93.6 7e-20
At1g79550 91.3 5e-19
ECU05g0320 59.3 2e-09
CE20568 30.8 0.59
SPCC1281.04 28.1 4.4
Hs4826686 27.7 5.5
> Hs4505763
Length=417
Score = 157 bits (396), Expect = 5e-39, Method: Compositional matrix adjust.
Identities = 73/96 (76%), Positives = 87/96 (90%), Gaps = 0/96 (0%)
Query 3 ANGNKVKADSKQVEAFRNSLTKLGDIFVNDAFGTAHRAHASMVGVNLPVKAAGLLMKKEL 62
A+GNKVKA+ ++EAFR SL+KLGD++VNDAFGTAHRAH+SMVGVNLP KA G LMKKEL
Sbjct 135 ASGNKVKAEPAKIEAFRASLSKLGDVYVNDAFGTAHRAHSSMVGVNLPQKAGGFLMKKEL 194
Query 63 DYFSKALENPQKPFLAILGGAKVKDKIQLIKNLLSK 98
+YF+KALE+P++PFLAILGGAKV DKIQLI N+L K
Sbjct 195 NYFAKALESPERPFLAILGGAKVADKIQLINNMLDK 230
> Hs20270259
Length=417
Score = 157 bits (396), Expect = 6e-39, Method: Compositional matrix adjust.
Identities = 73/95 (76%), Positives = 86/95 (90%), Gaps = 0/95 (0%)
Query 4 NGNKVKADSKQVEAFRNSLTKLGDIFVNDAFGTAHRAHASMVGVNLPVKAAGLLMKKELD 63
+G K+KA+ ++EAFR SL+KLGD++VNDAFGTAHRAH+SMVGVNLP KA+G LMKKELD
Sbjct 136 SGKKIKAEPDKIEAFRASLSKLGDVYVNDAFGTAHRAHSSMVGVNLPHKASGFLMKKELD 195
Query 64 YFSKALENPQKPFLAILGGAKVKDKIQLIKNLLSK 98
YF+KALENP +PFLAILGGAKV DKIQLIKN+L K
Sbjct 196 YFAKALENPVRPFLAILGGAKVADKIQLIKNMLDK 230
> SPBC14F5.04c
Length=414
Score = 155 bits (393), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 76/95 (80%), Positives = 83/95 (87%), Gaps = 1/95 (1%)
Query 4 NGNKVKADSKQVEAFRNSLTKLGDIFVNDAFGTAHRAHASMVGVNLPVKAAGLLMKKELD 63
+G KVKAD+ VEAFR SLT LGDIFVNDAFGTAHRAH+SMVGV+LP + +G LMKKELD
Sbjct 134 DGKKVKADASAVEAFRKSLTSLGDIFVNDAFGTAHRAHSSMVGVDLP-RVSGFLMKKELD 192
Query 64 YFSKALENPQKPFLAILGGAKVKDKIQLIKNLLSK 98
YFSKALENP +PFLAILGGAKV DKIQLI NLL K
Sbjct 193 YFSKALENPARPFLAILGGAKVADKIQLIDNLLDK 227
> CE13100
Length=417
Score = 146 bits (369), Expect = 8e-36, Method: Compositional matrix adjust.
Identities = 72/98 (73%), Positives = 81/98 (82%), Gaps = 0/98 (0%)
Query 1 TSANGNKVKADSKQVEAFRNSLTKLGDIFVNDAFGTAHRAHASMVGVNLPVKAAGLLMKK 60
A+G KVKADS V+ FR SLTKLGDI+VNDAFGTAHRAH+SMVGV +A+G L+K
Sbjct 132 VDASGAKVKADSAAVKKFRESLTKLGDIYVNDAFGTAHRAHSSMVGVEHSQRASGFLLKN 191
Query 61 ELDYFSKALENPQKPFLAILGGAKVKDKIQLIKNLLSK 98
EL YFSKAL+NP +PFLAILGGAKV DKIQLIKNLL K
Sbjct 192 ELSYFSKALDNPARPFLAILGGAKVADKIQLIKNLLDK 229
> YCR012w
Length=416
Score = 140 bits (352), Expect = 7e-34, Method: Compositional matrix adjust.
Identities = 64/95 (67%), Positives = 78/95 (82%), Gaps = 0/95 (0%)
Query 4 NGNKVKADSKQVEAFRNSLTKLGDIFVNDAFGTAHRAHASMVGVNLPVKAAGLLMKKELD 63
+G KVKA + V+ FR+ L+ L D+++NDAFGTAHRAH+SMVG +LP +AAG L++KEL
Sbjct 134 DGQKVKASKEDVQKFRHELSSLADVYINDAFGTAHRAHSSMVGFDLPQRAAGFLLEKELK 193
Query 64 YFSKALENPQKPFLAILGGAKVKDKIQLIKNLLSK 98
YF KALENP +PFLAILGGAKV DKIQLI NLL K
Sbjct 194 YFGKALENPTRPFLAILGGAKVADKIQLIDNLLDK 228
> 7295919
Length=415
Score = 135 bits (339), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 66/96 (68%), Positives = 78/96 (81%), Gaps = 0/96 (0%)
Query 3 ANGNKVKADSKQVEAFRNSLTKLGDIFVNDAFGTAHRAHASMVGVNLPVKAAGLLMKKEL 62
A+G KVKAD +V+ FR SL KLGD++VNDAFGTAHRAH+SM+G +AAGLL+ KEL
Sbjct 133 ASGGKVKADPAKVKEFRASLAKLGDVYVNDAFGTAHRAHSSMMGDGFEQRAAGLLLNKEL 192
Query 63 DYFSKALENPQKPFLAILGGAKVKDKIQLIKNLLSK 98
YFS+AL+ P PFLAILGGAKV DKIQLI+NLL K
Sbjct 193 KYFSQALDKPPNPFLAILGGAKVADKIQLIENLLDK 228
> 7295921
Length=389
Score = 123 bits (309), Expect = 8e-29, Method: Compositional matrix adjust.
Identities = 60/98 (61%), Positives = 74/98 (75%), Gaps = 0/98 (0%)
Query 4 NGNKVKADSKQVEAFRNSLTKLGDIFVNDAFGTAHRAHASMVGVNLPVKAAGLLMKKELD 63
N KVKAD +V+ FR L +LG+I+VNDAFGTAHR H+SM+G V+AAG L+ KEL+
Sbjct 107 NKKKVKADPAKVKEFRAKLAQLGEIYVNDAFGTAHRPHSSMMGDGYKVRAAGFLLDKELE 166
Query 64 YFSKALENPQKPFLAILGGAKVKDKIQLIKNLLSKSAA 101
YF+K L +P KPFLAILGGAK+ DKI LI NLL+ A
Sbjct 167 YFAKVLHDPAKPFLAILGGAKIADKIPLITNLLNNVTA 204
> At3g12780
Length=481
Score = 94.0 bits (232), Expect = 6e-20, Method: Compositional matrix adjust.
Identities = 48/93 (51%), Positives = 61/93 (65%), Gaps = 2/93 (2%)
Query 9 KADSKQVEAFRNSLTKLGDIFVNDAFGTAHRAHASMVGVN--LPVKAAGLLMKKELDYFS 66
K + K F L L D++VNDAFGTAHRAHAS GV L AG L++KELDY
Sbjct 200 KEEEKNDPEFAKKLASLADLYVNDAFGTAHRAHASTEGVTKFLKPSVAGFLLQKELDYLV 259
Query 67 KALENPQKPFLAILGGAKVKDKIQLIKNLLSKS 99
A+ NP++PF AI+GG+KV KI +I++LL K
Sbjct 260 GAVSNPKRPFAAIVGGSKVSSKIGVIESLLEKC 292
> At1g56190
Length=478
Score = 93.6 bits (231), Expect = 7e-20, Method: Compositional matrix adjust.
Identities = 48/93 (51%), Positives = 61/93 (65%), Gaps = 2/93 (2%)
Query 9 KADSKQVEAFRNSLTKLGDIFVNDAFGTAHRAHASMVGVN--LPVKAAGLLMKKELDYFS 66
K + K F L L D++VNDAFGTAHRAHAS GV L AG L++KELDY
Sbjct 197 KEEEKNEPDFAKKLASLADLYVNDAFGTAHRAHASTEGVTKFLKPSVAGFLLQKELDYLV 256
Query 67 KALENPQKPFLAILGGAKVKDKIQLIKNLLSKS 99
A+ NP++PF AI+GG+KV KI +I++LL K
Sbjct 257 GAVSNPKRPFAAIVGGSKVSSKIGVIESLLEKC 289
> At1g79550
Length=401
Score = 91.3 bits (225), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 47/82 (57%), Positives = 58/82 (70%), Gaps = 2/82 (2%)
Query 18 FRNSLTKLGDIFVNDAFGTAHRAHASMVGVN--LPVKAAGLLMKKELDYFSKALENPQKP 75
F L L D++VNDAFGTAHRAHAS GV L AG LM+KELDY A+ NP+KP
Sbjct 134 FAKKLAALADVYVNDAFGTAHRAHASTEGVAKFLKPSVAGFLMQKELDYLVGAVANPKKP 193
Query 76 FLAILGGAKVKDKIQLIKNLLS 97
F AI+GG+KV KI +I++LL+
Sbjct 194 FAAIVGGSKVSTKIGVIESLLN 215
> ECU05g0320
Length=388
Score = 59.3 bits (142), Expect = 2e-09, Method: Composition-based stats.
Identities = 35/92 (38%), Positives = 54/92 (58%), Gaps = 3/92 (3%)
Query 10 ADSKQVEAFRNSLTKLGDIFVNDAFGTAHRAHASMVGVNLPVKAAGLLMKKELDYFSKAL 69
D V+ +RN D+ V DAFG HR S+ LP +GLL++KEL++ + +
Sbjct 122 GDKDAVDQYRNVFVDNMDVAVIDAFGCLHRECGSIQRTGLP-SFSGLLVQKELNFTKEIM 180
Query 70 ENPQKPFLAILGGAKVKDKIQLIKNLLSKSAA 101
E ++ L ILGG KV DKI+L+++L K+ +
Sbjct 181 E--ERVDLMILGGKKVSDKIKLLESLGQKTGS 210
> CE20568
Length=305
Score = 30.8 bits (68), Expect = 0.59, Method: Compositional matrix adjust.
Identities = 20/70 (28%), Positives = 35/70 (50%), Gaps = 7/70 (10%)
Query 25 LGDIFVNDAFGTAHRAHASMVGVNLPVKAAGLLMKK-----ELDYFS-KALENPQKPFLA 78
+ DI +ND H H ++ G+NL +L+KK L+YF + +E+P + +
Sbjct 188 VHDIQLNDINLMVHCWHVALAGINLNSDHLNVLLKKWKENSSLEYFCAREMESPLEKTI- 246
Query 79 ILGGAKVKDK 88
IL G + +
Sbjct 247 ILEGTNSRQR 256
> SPCC1281.04
Length=333
Score = 28.1 bits (61), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 23/52 (44%), Gaps = 4/52 (7%)
Query 35 GTAHRAHASMVGVNLPVKAAGLLMKKELDYFSKALENPQKPFLAILGGAKVK 86
G+ + G+N P LL DYF K +N K FL++ GG K
Sbjct 42 GSNYWNAGEFYGINPPTANLDLLA----DYFEKYPKNADKVFLSVKGGTDFK 89
> Hs4826686
Length=740
Score = 27.7 bits (60), Expect = 5.5, Method: Composition-based stats.
Identities = 11/40 (27%), Positives = 22/40 (55%), Gaps = 0/40 (0%)
Query 65 FSKALENPQKPFLAILGGAKVKDKIQLIKNLLSKSAAAPA 104
F K ++NP+ L I+GG +D++ +++N + P
Sbjct 309 FKKYIDNPKLRELLIIGGVAARDQLSVLENGVDIVVGTPG 348
Lambda K H
0.313 0.127 0.339
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1195973986
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40