bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
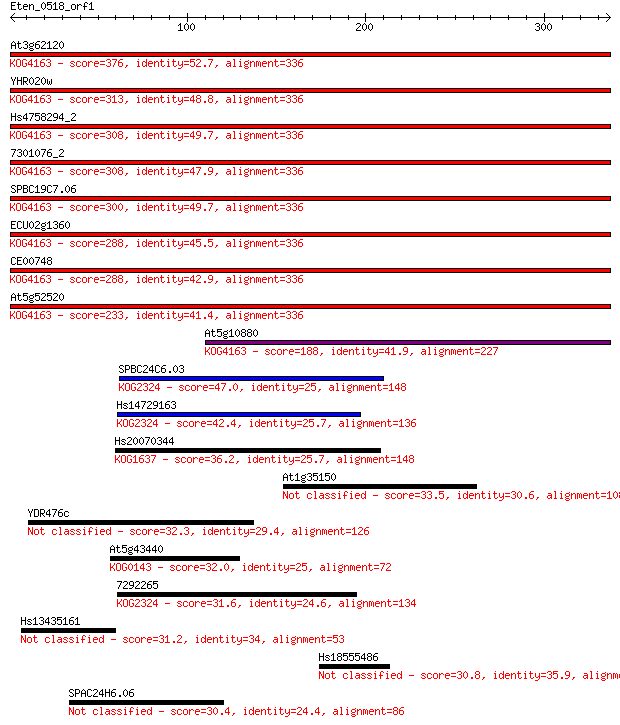
Query= Eten_0518_orf1
Length=336
Score E
Sequences producing significant alignments: (Bits) Value
At3g62120 376 4e-104
YHR020w 313 4e-85
Hs4758294_2 308 9e-84
7301076_2 308 2e-83
SPBC19C7.06 300 2e-81
ECU02g1360 288 1e-77
CE00748 288 1e-77
At5g52520 233 3e-61
At5g10880 188 2e-47
SPBC24C6.03 47.0 6e-05
Hs14729163 42.4 0.001
Hs20070344 36.2 0.11
At1g35150 33.5 0.70
YDR476c 32.3 1.6
At5g43440 32.0 1.8
7292265 31.6 2.4
Hs13435161 31.2 3.1
Hs18555486 30.8 4.2
SPAC24H6.06 30.4 6.0
> At3g62120
Length=530
Score = 376 bits (965), Expect = 4e-104, Method: Compositional matrix adjust.
Identities = 177/336 (52%), Positives = 236/336 (70%), Gaps = 12/336 (3%)
Query 1 TAHATEEEAYTLVLEILELYRQWYEDYLAVPVIKGEKSENEKFAGGKKTTTIEGIIPDTG 60
TA AT+ EA VL+ILELYR+ YE+YLAVPV+KG KSENEKFAGG TT++E IP+TG
Sbjct 207 TAFATKAEADEEVLQILELYRRIYEEYLAVPVVKGMKSENEKFAGGLYTTSVEAFIPNTG 266
Query 61 RGIQAATSHLLGQNFSRMFSIEFEDEKGAKQLVHQTSWGCTTRSLGVMIMTHGDDKGLVL 120
RG+Q ATSH LGQNF++MF I FE+EK ++V Q SW +TR++GVMIMTHGDDKGLVL
Sbjct 267 RGVQGATSHCLGQNFAKMFEINFENEKAETEMVWQNSWAYSTRTIGVMIMTHGDDKGLVL 326
Query 121 PPRVVAVQAVIIPIIFKETGGMEIVAKCRELERLLNAAGVRVKVDDRTNYTPGWKFNDWE 180
PP+V +VQ V+IP+ +K+ I C L AG+R + D R NY+PGWK++DWE
Sbjct 327 PPKVASVQVVVIPVPYKDANTQGIYDACTATASALCEAGIRAEEDLRDNYSPGWKYSDWE 386
Query 181 LKGVPLRLEIGPRDVESCQTRVVRRDTSEARNVPWAEAASTIPAMLETMQKDLFNKAKAK 240
+KGVPLR+EIGPRD+E+ Q R VRRD ++P + +LE +Q++++ AK K
Sbjct 387 MKGVPLRIEIGPRDLENDQVRTVRRDNGVKEDIPRGSLVEHVKELLEKIQQNMYEVAKQK 446
Query 241 FEASIEQITTFDEVMPALNRRHVVLAPWCEDPETETQIKRETQRLSEIQARENADGMTGA 300
EA ++++ T+DE + ALN + ++LAPWC++ E E +K T+ G TGA
Sbjct 447 REACVQEVKTWDEFIKALNEKKLILAPWCDEEEVERDVKARTK------------GETGA 494
Query 301 MKPLCIPFDQPPMPEGTRCFFTGRPAKRWCLFGRSY 336
K LC PFDQP +PEGT CF +G+PAK+W +GRSY
Sbjct 495 AKTLCSPFDQPELPEGTLCFASGKPAKKWTYWGRSY 530
> YHR020w
Length=688
Score = 313 bits (802), Expect = 4e-85, Method: Compositional matrix adjust.
Identities = 164/344 (47%), Positives = 220/344 (63%), Gaps = 8/344 (2%)
Query 1 TAHATEEEAYTLVLEILELYRQWYEDYLAVPVIKGEKSENEKFAGGKKTTTIEGIIPDTG 60
TAH T ++A VL+IL+ Y YE+ LAVPV+KG K+E EKFAGG TTT EG IP TG
Sbjct 345 TAHLTAKDAEEEVLQILDFYAGVYEELLAVPVVKGRKTEKEKFAGGDFTTTCEGYIPQTG 404
Query 61 RGIQAATSHLLGQNFSRMFSIEFEDEKGA---KQLVHQTSWGCTTRSLGVMIMTHGDDKG 117
RGIQ ATSH LGQNFS+MF++ E+ G+ K +Q SWG +TR +GVM+M H D+KG
Sbjct 405 RGIQGATSHHLGQNFSKMFNLSVENPLGSDHPKIFAYQNSWGLSTRVIGVMVMIHSDNKG 464
Query 118 LVLPPRVVAVQAVIIPI-IFKETGGME---IVAKCRELERLLNAAGVRVKVDDRTNYTPG 173
LV+PPRV Q+V+IP+ I K+T + I R +E L G+R D NYTPG
Sbjct 465 LVIPPRVSQFQSVVIPVGITKKTSEEQRKHIHETARSVESRLKKVGIRAFGDYNDNYTPG 524
Query 174 WKFNDWELKGVPLRLEIGPRDVESCQTRVVRRDTSEARNVPWAEAASTIPAMLETMQKDL 233
WKF+ +ELKG+P+R+E+GP+D+E Q VVRR+ S+ V + E + IP +LE MQ DL
Sbjct 525 WKFSQYELKGIPIRIELGPKDIEKNQVVVVRRNDSKKYVVSFDELEARIPEILEEMQGDL 584
Query 234 FNKAKAKFEASIEQITTFDEVMPALNRRHVVLAPWCEDPETETQIKRET-QRLSEIQARE 292
F KAK F+ + + +PALN+++V+LAPWC E E IK + ++ + E
Sbjct 585 FKKAKELFDTHRVIVNEWSGFVPALNKKNVILAPWCGVMECEEDIKESSAKKDDGEEFEE 644
Query 293 NADGMTGAMKPLCIPFDQPPMPEGTRCFFTGRPAKRWCLFGRSY 336
+ + K LCIPFDQP + EG +C R A +C+FGRSY
Sbjct 645 DDKAPSMGAKSLCIPFDQPVLNEGQKCIKCERIAVNYCMFGRSY 688
> Hs4758294_2
Length=499
Score = 308 bits (790), Expect = 9e-84, Method: Compositional matrix adjust.
Identities = 167/345 (48%), Positives = 213/345 (61%), Gaps = 15/345 (4%)
Query 1 TAHATEEEAYTLVLEILELYRQWYEDYLAVPVIKGEKSENEKFAGGKKTTTIEGIIPDTG 60
+A AT EEA VL+IL+LY Q YE+ LA+PV+KG K+E EKFAGG TTTIE I +G
Sbjct 161 SAFATMEEAAEEVLQILDLYAQVYEELLAIPVVKGRKTEKEKFAGGDYTTTIEAFISASG 220
Query 61 RGIQAATSHLLGQNFSRMFSIEFEDEK--GAKQLVHQTSWGCTTRSLGVMIMTHGDDKGL 118
R IQ TSH LGQNFS+MF I FED K G KQ +Q SWG TTR++GVM M HGD+ GL
Sbjct 221 RAIQGGTSHHLGQNFSKMFEIVFEDPKIPGEKQFAYQNSWGLTTRTIGVMTMVHGDNMGL 280
Query 119 VLPPRVVAVQAVIIPI----IFKETGGMEIVAKCRELERLLNAAGVRVKVDDRTNYTPGW 174
VLPPRV VQ VIIP E ++AKC + R L + +RV+ D R NY+PGW
Sbjct 281 VLPPRVACVQVVIIPCGITNALSEEDKEALIAKCNDYRRRLLSVNIRVRADLRDNYSPGW 340
Query 175 KFNDWELKGVPLRLEIGPRDVESCQTRVVRRDTSEARNVPWAEAASTIPAMLETMQKDLF 234
KFN WELKGVP+RLE+GPRD++SCQ VRRDT E V EA + + A+LE +Q LF
Sbjct 341 KFNHWELKGVPIRLEVGPRDMKSCQFVAVRRDTGEKLTVAENEAETKLQAILEDIQVTLF 400
Query 235 NKAKAKFEASIEQITTFDEVMPALNRRHVVLAPWCEDPETETQIKRETQRLSEIQARENA 294
+A + + T ++ L+ +V P+C + + E IK+ T R +++ +
Sbjct 401 TRASEDLKTHMVVANTMEDFQKILDSGKIVQIPFCGEIDCEDWIKKTTARDQDLEPGAPS 460
Query 295 DGMTGAMKPLCIPFDQPPMPE---GTRCFFTGRPAKRWCLFGRSY 336
G K LCIPF P+ E G +C PAK + LFGRSY
Sbjct 461 MG----AKSLCIPFK--PLCELQPGAKCVCGKNPAKYYTLFGRSY 499
> 7301076_2
Length=586
Score = 308 bits (788), Expect = 2e-83, Method: Compositional matrix adjust.
Identities = 161/345 (46%), Positives = 223/345 (64%), Gaps = 14/345 (4%)
Query 1 TAHATEEEAYTLVLEILELYRQWYEDYLAVPVIKGEKSENEKFAGGKKTTTIEGIIPDTG 60
TA A +EEA VL+IL+LY Y LA+PV+KG K+E EKFAGG TTT+E I +G
Sbjct 247 TAFADKEEAAKEVLDILDLYALVYTHLLAIPVVKGRKTEKEKFAGGDYTTTVEAFISASG 306
Query 61 RGIQAATSHLLGQNFSRMFSIEFED-EKGAKQLVHQTSWGCTTRSLGVMIMTHGDDKGLV 119
R IQ ATSH LGQNFS+MF I +ED E K+ V+Q SWG TTR++GVMIM H D++GLV
Sbjct 307 RAIQGATSHHLGQNFSKMFEIVYEDPETQQKKYVYQNSWGITTRTIGVMIMVHADNQGLV 366
Query 120 LPPRVVAVQAVIIP----IIFKETGGMEIVAKCRELERLLNAAGVRVKVDDRTNYTPGWK 175
LPP V +QA+++P + K+ +++ C+ LE+ L GVR + D R NY+PGWK
Sbjct 367 LPPHVACIQAIVVPCGITVNTKDDERAQLLDACKALEKRLVGGGVRCEGDYRDNYSPGWK 426
Query 176 FNDWELKGVPLRLEIGPRDVESCQTRVVRRDTSEARNVPWAEAASTIPAMLETMQKDLFN 235
FN WELKGVPLRLE+GP+D+++ Q VRRDT E +P A+ IPA+LET+ + + N
Sbjct 427 FNHWELKGVPLRLEVGPKDLKAQQLVAVRRDTVEKITIPLADVEKKIPALLETIHESMLN 486
Query 236 KAKAKFEASIEQITTFDEVMPALNRRHVVLAPWCEDPETETQIKRETQRLSEIQARENAD 295
KA+ + +++T + + L +++++LAP+C + E +IK ++ R E + A
Sbjct 487 KAQEDMTSHTKKVTNWTDFCGFLEQKNILLAPFCGEISCEDKIKADSARGEEAEPGAPAM 546
Query 296 GMTGAMKPLCIPFDQP-PMPEGTRCF---FTGRPAKRWCLFGRSY 336
G K LCIPFDQP P+ +C T +P K + LFGRSY
Sbjct 547 G----AKSLCIPFDQPAPIAASDKCINPSCTNKP-KFYTLFGRSY 586
> SPBC19C7.06
Length=716
Score = 300 bits (769), Expect = 2e-81, Method: Compositional matrix adjust.
Identities = 167/353 (47%), Positives = 215/353 (60%), Gaps = 18/353 (5%)
Query 1 TAHATEEEAYTLVLEILELYRQWYEDYLAVPVIKGEKSENEKFAGGKKTTTIEGIIPDTG 60
TAH T E A V +IL+LY + Y D LAVPVIKG KSENEKFAGG TTT+EG IP TG
Sbjct 365 TAHMTLEGATEEVHQILDLYARIYTDLLAVPVIKGVKSENEKFAGGMFTTTVEGYIPTTG 424
Query 61 RGIQAATSHLLGQNFSRMFSIEFEDEKG--------AKQLVHQTSWGCTTRSLGVMIMTH 112
RGIQ ATSH LGQNFS+MF+I ED K V Q SWG +TR++GV +M H
Sbjct 425 RGIQGATSHCLGQNFSKMFNIVVEDPNAEIGPTGERPKLFVWQNSWGLSTRTIGVAVMVH 484
Query 113 GDDKGLVLPPRVVAVQAVIIP--IIFKETGG--MEIVAKCRELERLLNAAGVRVKVDDRT 168
GDDKGL LPP + VQ+V++P I K T EI C +L LNAA +R + D R
Sbjct 485 GDDKGLKLPPAIALVQSVVVPCGITNKTTDQERNEIEGFCSKLADRLNAADIRTEADLRA 544
Query 169 NYTPGWKFNDWELKGVPLRLEIGPRDVESCQTRVVRRDTSEARNVPWAEAASTIPAMLET 228
YTPG+KF+ WE+KGVPLRLE GP D + Q VRRDT E VP + +L
Sbjct 545 -YTPGYKFSHWEMKGVPLRLEYGPNDAKKNQVTAVRRDTFEKIPVPLNNLEKGVSDLLAK 603
Query 229 MQKDLFNKAKAKFEASIEQITTFDEVMPALNRRHVVLAPWCEDPETETQIKRETQRLSEI 288
+Q +++ AKA+ +A + ++ + + +PALN++++V+ PWC E E +IK+ + R
Sbjct 604 IQTNMYETAKAERDAHVVKVKEWADFVPALNKKNIVMIPWCNTTECEKEIKKNSARQVNG 663
Query 289 QARENADGMTGAMKPLCIPFDQPP----MPEG-TRCFFTGRPAKRWCLFGRSY 336
E+ + K LCIP +QP + EG T+C G AK W LFGRSY
Sbjct 664 DEPEDEKAPSMGAKSLCIPLEQPSGEDAIIEGTTKCAGCGNLAKVWGLFGRSY 716
> ECU02g1360
Length=520
Score = 288 bits (736), Expect = 1e-77, Method: Compositional matrix adjust.
Identities = 153/348 (43%), Positives = 212/348 (60%), Gaps = 24/348 (6%)
Query 1 TAHATEEEAYTLVLEILELYRQWYEDYLAVPVIKGEKSENEKFAGGKKTTTIEGIIPDTG 60
TA T +E+ VL IL+LY Q Y + LAVPVIKG KSENEKF G TT+IE IP +G
Sbjct 185 TAFLTRKESDEEVLAILDLYSQIYSELLAVPVIKGRKSENEKFGGADYTTSIEAFIPGSG 244
Query 61 RGIQAATSHLLGQNFSRMFSIEFEDEKGAKQ--LVHQTSWGCTTRSLGVMIMTHGDDKGL 118
RG+QAATSH LGQNFSRMF I+ + ++G++ V+Q SWG TTRS+G+ M H D+ GL
Sbjct 245 RGVQAATSHSLGQNFSRMFDIKADTDEGSESSSFVYQNSWGITTRSIGIAAMIHSDNLGL 304
Query 119 VLPPRVVAVQAVIIPIIFKETGGMEIVAKCRELERL----------LNAAGVRVKVDDRT 168
VLPPRV Q VI+P G+ + + E L L +GVRV +DDR+
Sbjct 305 VLPPRVAMTQVVIVPC------GITTASSKDDTESLRAYINGVCVQLKNSGVRVHLDDRS 358
Query 169 NYTPGWKFNDWELKGVPLRLEIGPRDVESCQTRVVRRDTSEARNVPWAEAASTIPAMLET 228
N T G+KFN WE++GVPLRLEIG +D+ S + +VRRDT + V A T+ ++T
Sbjct 359 NVTAGFKFNHWEIRGVPLRLEIGFKDMASSEACLVRRDTRAKKQVSVEGIAHTVMEEIDT 418
Query 229 MQKDLFNKAKAKFEASIEQITTFDEVMPALNRRHVVLAPWCEDPETETQIKRETQRLSEI 288
M D+ +A ++ ++ I + +F+E M AL+ +++++APWC E E +IK + R
Sbjct 419 MHNDMLARATSERDSRISYVKSFEEFMSALDNKNIIMAPWCGISECEIEIKSRSTR---- 474
Query 289 QARENADGMTGAMKPLCIPFDQPPMPEGTRCFFTGRPAKRWCLFGRSY 336
A +D ++ K LCIP+ P +G +C A + LFGRSY
Sbjct 475 -ADPRSDVVSTGAKTLCIPYGSKPC-DGMKCINCNSQAVHYTLFGRSY 520
> CE00748
Length=581
Score = 288 bits (736), Expect = 1e-77, Method: Compositional matrix adjust.
Identities = 144/340 (42%), Positives = 204/340 (60%), Gaps = 11/340 (3%)
Query 1 TAHATEEEAYTLVLEILELYRQWYEDYLAVPVIKGEKSENEKFAGGKKTTTIEGIIPDTG 60
TA A +A V +IL+LY Y D LA+PV+KG KSE EKFAGG TTT+E + G
Sbjct 249 TAFANPADAEKEVFQILDLYAGVYTDLLAIPVVKGRKSEKEKFAGGDFTTTVEAYVACNG 308
Query 61 RGIQAATSHLLGQNFSRMFSIEFED--EKGAKQLVHQTSWGCTTRSLGVMIMTHGDDKGL 118
RGIQ ATSH LGQNFS+MF I +ED ++G + Q SWG +TR++G M+M HGDDKGL
Sbjct 309 RGIQGATSHHLGQNFSKMFDISYEDPAKEGERAFAWQNSWGLSTRTIGAMVMIHGDDKGL 368
Query 119 VLPPRVVAVQAVIIPIIFKETGGMEIVAKCRELERLLNAAGVRVKVDDRTNYTPGWKFND 178
VLPPRV AVQ +++P+ FK + ++ + L A GV+ + D R NY GWKFN
Sbjct 369 VLPPRVAAVQVIVVPVGFKTENKQSLFDAVDKVTKDLVADGVKAEHDLRENYNAGWKFNH 428
Query 179 WELKGVPLRLEIGPRDVESCQTRVVRRDTSEARNVPWAEAASTIPAMLETMQKDLFNKAK 238
WELKGVP+R E+GP D+ Q V R E + + + T+ ++L+ + ++NK
Sbjct 429 WELKGVPIRFEVGPNDLAKNQVTAVIRHNGEKKQLSLEGLSKTVKSLLDEIHTAMYNKVL 488
Query 239 AKFEASIEQITTFDEVMPALNRRHVVLAPWCEDPETETQIKRETQRLSEIQARENADGMT 298
A+ ++ ++ T +E L+ + ++L+P+C P+ E +IK+ + RE+ +G
Sbjct 489 AQRDSHLKSTTCIEEFKKLLDEKCIILSPFCGRPDCEEEIKKAS-------TREDGEGAQ 541
Query 299 GAMKPLCIPFDQPPMPEGTRCFFTG--RPAKRWCLFGRSY 336
K LCIP DQP ++C F AK + LFGRSY
Sbjct 542 MGAKTLCIPLDQPKQELPSKCLFPSCTYNAKAFALFGRSY 581
> At5g52520
Length=543
Score = 233 bits (595), Expect = 3e-61, Method: Compositional matrix adjust.
Identities = 139/343 (40%), Positives = 195/343 (56%), Gaps = 28/343 (8%)
Query 1 TAHATEEEAYTLVLEILELYRQWYEDYLAVPVIKGEKSENEKFAGGKKTTTIEGIIPDTG 60
TAHAT EEA +++E+Y ++ + A+PVI G KS+ E FAG T TIE ++ D
Sbjct 222 TAHATPEEAEKEAKQMIEIYTRFAFEQTAIPVIPGRKSKLETFAGADITYTIEAMMGDR- 280
Query 61 RGIQAATSHLLGQNFSRMFSIEFEDEKGAKQLVHQTSWGCTTRSLGVMIMTHGDDKGLVL 120
+ +QA TSH LGQNFSR F +F DE G +Q V QTSW +TR +G +IMTHGDD GL+L
Sbjct 281 KALQAGTSHNLGQNFSRAFGTQFADENGERQHVWQTSWAVSTRFVGGIIMTHGDDTGLML 340
Query 121 PPRVVAVQAVIIPIIFKETGGMEIVAKCRELERLLNAAGVRVKVDDRTNYTPGWKFNDWE 180
PP++ +Q VI+PI K+T +++ ++ L AGVRVK+DD TPGWKFN WE
Sbjct 341 PPKIAPIQVVIVPIWKKDTEKTGVLSAASSVKEALQTAGVRVKLDDTDQRTPGWKFNFWE 400
Query 181 LKGVPLRLEIGPRDVESCQTRVVRRDT--SEARNVPWAEAASTIPAM----LETMQKDLF 234
+KG+PLR+EIGPRDV S V RRD + + ST+ A L+ +Q L
Sbjct 401 MKGIPLRIEIGPRDVSSNSVVVSRRDVPGKAGKVFGISMEPSTLVAYVKEKLDEIQTSLL 460
Query 235 NKAKAKFEASIEQITTFDEVMPALNRRHVVLAPWCEDPETETQIKRETQRLSEIQARENA 294
KA + +++I + ++ E+ A++ PW E ++K E
Sbjct 461 EKALSFRDSNIVDVNSYAELKDAISSGKWARGPWSASDADEQRVKEE------------- 507
Query 295 DGMTGAMKPLCIPFDQPPMPEGTR-CFFTGRPAKRWCLFGRSY 336
TGA C PF+Q +GT+ C TG PA+ +F +SY
Sbjct 508 ---TGATI-RCFPFEQ---TQGTKTCLMTGNPAEEVAIFAKSY 543
> At5g10880
Length=309
Score = 188 bits (477), Expect = 2e-47, Method: Compositional matrix adjust.
Identities = 95/228 (41%), Positives = 136/228 (59%), Gaps = 8/228 (3%)
Query 110 MTHGDDKGLVLPPRVVAVQAVIIPIIFKETGGM-EIVAKCRELERLLNAAGVRVKVDDRT 168
MTHGDDKGLV PP+V VQ V+I + K E+ C +E L AG+R + D R
Sbjct 89 MTHGDDKGLVFPPKVAPVQVVVIHVPIKGAADYQELCDACEAVESTLLGAGIRAEADIRD 148
Query 169 NYTPGWKFNDWELKGVPLRLEIGPRDVESCQTRVVRRDTSEARNVPWAEAASTIPAMLET 228
NY+ GWK+ D EL GVPLR+E GPRD+ + Q R+V RD +V + + +LE
Sbjct 149 NYSCGWKYADQELTGVPLRIETGPRDLANDQVRIVTRDNGAKMDVKRGDLIEQVKDLLEK 208
Query 229 MQKDLFNKAKAKFEASIEQITTFDEVMPALNRRHVVLAPWCEDPETETQIKRETQRLSEI 288
+Q +L++ AK K E +++ T+DE + AL+++ ++LAPWC+ E E +KR T+
Sbjct 209 IQSNLYDVAKRKVEECTQKVETWDEFVEALSQKKLILAPWCDKVEVEKDVKRRTR----- 263
Query 289 QARENADGMTGAMKPLCIPFDQPPMPEGTRCFFTGRPAKRWCLFGRSY 336
+ G G K LC P +QP + E T CF +G+PAK+W +GRSY
Sbjct 264 --GDETGGGGGGAKTLCTPLEQPELGEETLCFASGKPAKKWSYWGRSY 309
> SPBC24C6.03
Length=425
Score = 47.0 bits (110), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 37/150 (24%), Positives = 64/150 (42%), Gaps = 8/150 (5%)
Query 62 GIQAATSHLLGQNFSRMFSIEFEDEKGAKQLVHQTSWGC-TTRSLGVMIMTHGDDKGLVL 120
I+ + LG+ +S F+ E K ++++H +G +R + + D KGLV
Sbjct 264 AIEVGHAFYLGKIYSSKFNATVE-VKNKQEVLHMGCYGIGVSRLIAAVAHVTKDAKGLVW 322
Query 121 PPRVVAVQAVIIPIIFKETGGMEIVAKCRELERLLNAAGV-RVKVDDRTNYTPGWKFNDW 179
P + + +++P E V + N G V ++DR N G+K D
Sbjct 323 PSSIAPWKVLVVPTSDNHIQSAETV-----YDATANVVGFDNVLLEDRQNRAFGYKMRDA 377
Query 180 ELKGVPLRLEIGPRDVESCQTRVVRRDTSE 209
EL G P + +G R E ++ R + E
Sbjct 378 ELIGYPFVIVVGSRFQEEGICEIIVRSSGE 407
> Hs14729163
Length=402
Score = 42.4 bits (98), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 35/140 (25%), Positives = 59/140 (42%), Gaps = 4/140 (2%)
Query 61 RGIQAATSHLLGQNFSRMFSIEFEDEKGAKQLVHQTSWGC-TTRSLGVMIMTHGDDKGLV 119
+GI+ + LG +S +F+ +F + G L +G TR L I + +
Sbjct 230 KGIEVGHTFYLGTKYSSIFNAQFTNVCGKPTLAEMGCYGLGVTRILAAAIEVLSTEDCVR 289
Query 120 LPPRVVAVQAVIIPII--FKETGGMEIVAKCRE-LERLLNAAGVRVKVDDRTNYTPGWKF 176
P + QA +IP KE E++ + + + + V +DDRT+ T G +
Sbjct 290 WPSLLAPYQACLIPPKKGSKEQAASELIGQLYDHITEAVPQLHGEVLLDDRTHLTIGNRL 349
Query 177 NDWELKGVPLRLEIGPRDVE 196
D G P + G R +E
Sbjct 350 KDANKFGYPFVIIAGKRALE 369
> Hs20070344
Length=718
Score = 36.2 bits (82), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 38/151 (25%), Positives = 63/151 (41%), Gaps = 14/151 (9%)
Query 60 GRGIQAATSHLLGQNFSRMFSIEFEDEKGAKQ---LVHQTSWGCTTRSLGVMIMTHGDDK 116
GR Q T L Q R F ++++ + GA + L+H+ G R LGV+ + G
Sbjct 549 GRPHQCGTIQLDFQLPLR-FDLQYKGQAGALERPVLIHRAVLGSVERLLGVLAESCGGKW 607
Query 117 GLVLPPRVVAVQAVIIPIIFKETGGMEIVAKCRELERLLNAAGVRVKVDDRTNYTPGWKF 176
L L P Q V+IP+ G E +E ++ L AAG+ +D + T +
Sbjct 608 PLWLSP----FQVVVIPV------GSEQEEYAKEAQQSLRAAGLVSDLDADSGLTLSRRI 657
Query 177 NDWELKGVPLRLEIGPRDVESCQTRVVRRDT 207
+L + +G ++ + RD
Sbjct 658 RRAQLAHYNFQFVVGQKEQSKRTVNIRTRDN 688
> At1g35150
Length=459
Score = 33.5 bits (75), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 33/133 (24%), Positives = 47/133 (35%), Gaps = 25/133 (18%)
Query 154 LLNAAGVRVKVDDRTNYTPGWKFNDWELKGVPLRLEIGPRDVESCQTR------------ 201
LL G+ + D + Y +NDW L+ G D +C TR
Sbjct 80 LLVEKGLATRHDKKGGYLATSGYNDWRNLSKRLKEHKGSHDHITCMTRRAELESRLQKNK 139
Query 202 VVRRDTSEA---RNVPWAEAASTIPAMLETMQKDLF----------NKAKAKFEASIEQI 248
+ + EA N+ W E I A+++T K+ F + IE I
Sbjct 140 TIDKHAQEAINKDNIHWREVLLRIIALVKTHAKNNLAFRGKNEKVGQDRNGNFLSFIEMI 199
Query 249 TTFDEVMPALNRR 261
FD VM RR
Sbjct 200 AEFDVVMREHIRR 212
> YDR476c
Length=224
Score = 32.3 bits (72), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 37/146 (25%), Positives = 59/146 (40%), Gaps = 29/146 (19%)
Query 11 TLVLEILELYRQWYEDYLAV----PVIKGEK------------SENEKFAGGKKTTTIEG 54
+L++ I + ++ ED LAV P+ K K + KF + IE
Sbjct 4 SLIVSINDTHKLGLEDCLAVFGHVPITKAVKHVRLTEIDTQTSTFTLKFLHTETGQNIEK 63
Query 55 II----PDTGRGIQAATSHLLGQNFSRMFSIEFEDEKGAKQLVHQTSWGCTTRSLGVMIM 110
II DTG + AT + Q F++MF I E K + + + CT L +++
Sbjct 64 IIYFIDNDTGNDTRTATG--IKQIFNKMFRIAAEKRKLSLIQIDTVEYPCTLVDLLILV- 120
Query 111 THGDDKGLVLPPRVVAVQAVIIPIIF 136
G+ LPP + + I F
Sbjct 121 ------GVALPPLCYLYRPALHAIFF 140
> At5g43440
Length=365
Score = 32.0 bits (71), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 18/72 (25%), Positives = 34/72 (47%), Gaps = 7/72 (9%)
Query 57 PDTGRGIQAATSHLLGQNFSRMFSIEFEDEKGAKQLVHQTSWGCTTRSLGVMIMTHGDDK 116
PD GI T + +I +D+ G Q++HQ SW T G ++++ GD
Sbjct 229 PDLTLGISKHTD-------NSFITILLQDQIGGLQVLHQDSWVDVTPVPGALVISIGDFM 281
Query 117 GLVLPPRVVAVQ 128
L+ + ++++
Sbjct 282 QLITNDKFLSME 293
> 7292265
Length=458
Score = 31.6 bits (70), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 33/139 (23%), Positives = 56/139 (40%), Gaps = 7/139 (5%)
Query 61 RGIQAATSHLLGQNFSRMFSIEFEDEKGAKQLVHQTSWGC-TTRSLGVMIMTHGDDKGLV 119
RG++ A + LLG +S+ F + G Q + +G TR + + D L
Sbjct 262 RGVEVAHTFLLGDKYSKPLGATFLNTTGKPQSLVMGCYGIGITRVIAAALEVLSSDHELR 321
Query 120 LPPRVVAVQAVIIPIIFKETGGMEIVAKCRELERLLNAAGV----RVKVDDRTNYTPGWK 175
P + +I K+ + A+ E E LLN + + DDR T G +
Sbjct 322 WPKLLAPYDVCLIGP--KQGSKEQPEAEVIENELLLNVGEICGHQELLHDDRKELTIGKR 379
Query 176 FNDWELKGVPLRLEIGPRD 194
+ + G PL + +G +
Sbjct 380 LLEAKRLGHPLTIVVGAKS 398
> Hs13435161
Length=2559
Score = 31.2 bits (69), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 18/53 (33%), Positives = 27/53 (50%), Gaps = 0/53 (0%)
Query 7 EEAYTLVLEILELYRQWYEDYLAVPVIKGEKSENEKFAGGKKTTTIEGIIPDT 59
E+ + V E+ L RQW D LAV + GE E + G K+ + +PD+
Sbjct 855 EQLFAEVTEVAALRRQWLRDALAVYRMFGEVHACELWIGEKEQWLLSMRVPDS 907
> Hs18555486
Length=256
Score = 30.8 bits (68), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 23/40 (57%), Gaps = 3/40 (7%)
Query 174 WKFNDWELKGVPLRLEIGPRDV-ESCQTRVVRRDTSEARN 212
W+ W L +P R+ +GP+DV + CQ + R D+ R+
Sbjct 134 WELEGWTLANIPYRMPMGPQDVLDICQIK--RCDSVYTRS 171
> SPAC24H6.06
Length=668
Score = 30.4 bits (67), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 21/86 (24%), Positives = 35/86 (40%), Gaps = 6/86 (6%)
Query 34 KGEKSENEKFAGGKKTTTIEGIIPDTGRGIQAATSHLLGQNFSRMFSIEFEDEKGAKQLV 93
K S +FA T ++ P R +++ HL F ++ + KQ+V
Sbjct 179 KDGNSLTHEFANDSVTEMVQDYTPSCSRDVKSLLDHLYNSYFYQLLMTKTPVVFYVKQMV 238
Query 94 HQTSWGCTTRSLGVMIMTHGDDKGLV 119
+ TR L V + H ++K LV
Sbjct 239 GK------TRQLAVEVHNHVEEKALV 258
Lambda K H
0.319 0.134 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7988630762
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40