bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0611_orf1
Length=151
Score E
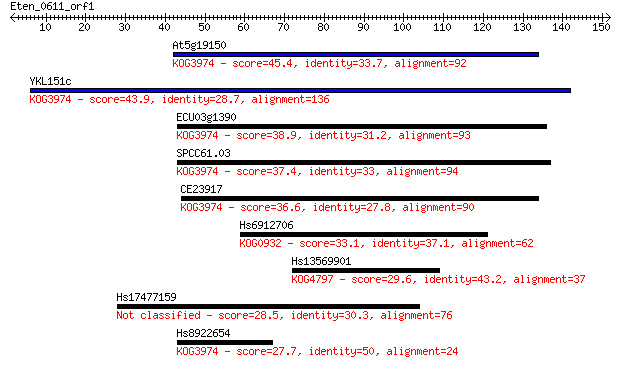
Sequences producing significant alignments: (Bits) Value
At5g19150 45.4 4e-05
YKL151c 43.9 1e-04
ECU03g1390 38.9 0.004
SPCC61.03 37.4 0.010
CE23917 36.6 0.020
Hs6912706 33.1 0.21
Hs13569901 29.6 2.1
Hs17477159 28.5 4.6
Hs8922654 27.7 9.0
> At5g19150
Length=382
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 31/92 (33%), Positives = 48/92 (52%), Gaps = 17/92 (18%)
Query 42 FGSPKRSGGQGDVLCGVLAEALVWSERGRALELKQVEEETAKLGLVVPFGGRPSVPYLFL 101
+GSP+R GGQGD+L G +A L W+++ ++ + + E A LG +
Sbjct 277 YGSPRRCGGQGDILSGGVAVFLSWAQQLKS-DPESPSENPAILGCI-------------- 321
Query 102 MHAASLITRQAAFLAYKHHGRSMTAEHILDRL 133
AAS + R+AA LA+ H RS I++ L
Sbjct 322 --AASGLLRKAASLAFTKHKRSTLTSDIIECL 351
> YKL151c
Length=337
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 39/137 (28%), Positives = 62/137 (45%), Gaps = 16/137 (11%)
Query 6 VFAKGRVDLLGAFDAKEDLEDCPRVYMAVAGLDPRYFGSPKRSGGQGDVLCGVLAEALVW 65
V KG+ D + + D+++D+ L GS KR GGQGD L G ++ L +
Sbjct 215 VVEKGQSDKIFSPDSEKDM------------LTNSEEGSNKRVGGQGDTLTGAISCMLAF 262
Query 66 SERGRALELKQVEEETAKLGLVVPFGGRPSVPYLFL-MHAASLITRQAAFLAYKHHGRSM 124
S ++ + +EE + P + V Y L +A ITR+ + L +K GR+M
Sbjct 263 SRAMYDFKICE-QEEKGESSNDKPL--KNWVDYAMLSCYAGCTITRECSRLGFKAKGRAM 319
Query 125 TAEHILDRLSAAAAIAF 141
+ DR+ A F
Sbjct 320 QTTDLNDRVGEVFAKLF 336
> ECU03g1390
Length=266
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 29/93 (31%), Positives = 40/93 (43%), Gaps = 23/93 (24%)
Query 43 GSPKRSGGQGDVLCGVLAEALVWSERGRALELKQVEEETAKLGLVVPFGGRPSVPYLFLM 102
G PKR GGQGD+L G +A +LV + P G+ + L
Sbjct 190 GCPKRIGGQGDILAGTIA-SLVSKCKA-------------------PVAGQDVFGSVVL- 228
Query 103 HAASLITRQAAFLAYKHHGRSMTAEHILDRLSA 135
++ R+A LAYK H RS+ IL+ L
Sbjct 229 --GCIMVRRAGRLAYKRHQRSLITRDILEELKT 259
> SPCC61.03
Length=327
Score = 37.4 bits (85), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 31/95 (32%), Positives = 43/95 (45%), Gaps = 14/95 (14%)
Query 43 GSPKRSGGQGDVLCGVLAEALVWSERGRALELKQVEEETAKLGLVVPFGGRPSVPYLFL- 101
G KR GGQGD+L G+LA L W A K+ + E G + LFL
Sbjct 228 GGLKRCGGQGDILTGILATFLAWR---HAYLSKEWDTE----------GNMDAKECLFLA 274
Query 102 MHAASLITRQAAFLAYKHHGRSMTAEHILDRLSAA 136
AS TR + LA+K GR+ + ++ + A
Sbjct 275 AFGASACTRWCSRLAFKECGRATQSTDLVRHVGKA 309
> CE23917
Length=307
Score = 36.6 bits (83), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 25/90 (27%), Positives = 36/90 (40%), Gaps = 20/90 (22%)
Query 44 SPKRSGGQGDVLCGVLAEALVWSERGRALELKQVEEETAKLGLVVPFGGRPSVPYLFLMH 103
S +R GGQGDV G L L W+++ + E
Sbjct 215 SLRRCGGQGDVTAGSLGLFLYWAKKNLGDDWTSAHHEAGI-------------------- 254
Query 104 AASLITRQAAFLAYKHHGRSMTAEHILDRL 133
A+S + R A A++ HGRSM +LD +
Sbjct 255 ASSWLVRTAGRRAFEKHGRSMNTPLLLDEI 284
> Hs6912706
Length=1056
Score = 33.1 bits (74), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 23/64 (35%), Positives = 31/64 (48%), Gaps = 2/64 (3%)
Query 59 LAEALVWSERGRALELKQVEEETAKLGLVVPF--GGRPSVPYLFLMHAASLITRQAAFLA 116
L +AL WS R LE EE+TA+ P G+ S P+L L ++ T + LA
Sbjct 725 LLKALYWSIRSEKLEWAVDEEDTARPEKAQPSLPAGKMSKPFLQLAQDPTVPTYKQGILA 784
Query 117 YKHH 120
K H
Sbjct 785 RKMH 788
> Hs13569901
Length=395
Score = 29.6 bits (65), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 16/37 (43%), Positives = 20/37 (54%), Gaps = 4/37 (10%)
Query 72 LELKQVEEETAKLGLVVPFGGRPSVPYLFLMHAASLI 108
+EL EE +G V P RPS P L+ H ASL+
Sbjct 244 MELGAPEE----MGQVPPLDSRPSSPALYFTHDASLV 276
> Hs17477159
Length=566
Score = 28.5 bits (62), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 23/78 (29%), Positives = 29/78 (37%), Gaps = 7/78 (8%)
Query 28 PRVYMAVAGLDPRYFGSPKRSGGQGDVLCGVLAEALVWSERGRALELKQVEEETAKLGLV 87
P+ + A A F SP+ G LC E VW R E V E + +
Sbjct 127 PQAFRAAA-----QFSSPRVGQGSATALCWSPTENWVWGVGARGAESPAVSEGPSTAAPL 181
Query 88 VPFGGRP--SVPYLFLMH 103
P G P S P LF +
Sbjct 182 RPRPGDPHSSSPMLFRLR 199
> Hs8922654
Length=390
Score = 27.7 bits (60), Expect = 9.0, Method: Compositional matrix adjust.
Identities = 12/24 (50%), Positives = 16/24 (66%), Gaps = 0/24 (0%)
Query 43 GSPKRSGGQGDVLCGVLAEALVWS 66
GS +R GGQGD+L G L + W+
Sbjct 265 GSSRRCGGQGDLLSGSLGVLVHWA 288
Lambda K H
0.322 0.137 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1888713112
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40