bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
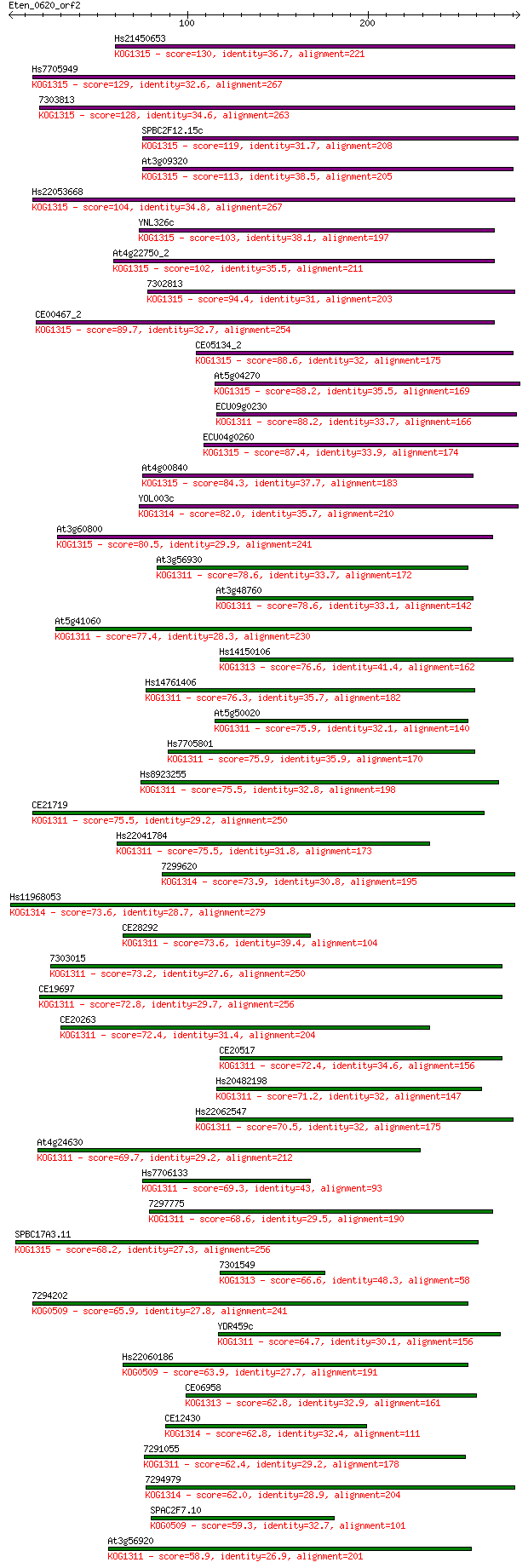
Query= Eten_0620_orf2
Length=283
Score E
Sequences producing significant alignments: (Bits) Value
Hs21450653 130 3e-30
Hs7705949 129 5e-30
7303813 128 1e-29
SPBC2F12.15c 119 9e-27
At3g09320 113 4e-25
Hs22053668 104 3e-22
YNL326c 103 4e-22
At4g22750_2 102 1e-21
7302813 94.4 3e-19
CE00467_2 89.7 5e-18
CE05134_2 88.6 1e-17
At5g04270 88.2 2e-17
ECU09g0230 88.2 2e-17
ECU04g0260 87.4 3e-17
At4g00840 84.3 3e-16
YOL003c 82.0 1e-15
At3g60800 80.5 4e-15
At3g56930 78.6 1e-14
At3g48760 78.6 1e-14
At5g41060 77.4 3e-14
Hs14150106 76.6 5e-14
Hs14761406 76.3 6e-14
At5g50020 75.9 8e-14
Hs7705801 75.9 8e-14
Hs8923255 75.5 1e-13
CE21719 75.5 1e-13
Hs22041784 75.5 1e-13
7299620 73.9 4e-13
Hs11968053 73.6 4e-13
CE28292 73.6 5e-13
7303015 73.2 5e-13
CE19697 72.8 7e-13
CE20263 72.4 9e-13
CE20517 72.4 1e-12
Hs20482198 71.2 3e-12
Hs22062547 70.5 4e-12
At4g24630 69.7 6e-12
Hs7706133 69.3 8e-12
7297775 68.6 1e-11
SPBC17A3.11 68.2 2e-11
7301549 66.6 6e-11
7294202 65.9 1e-10
YDR459c 64.7 2e-10
Hs22060186 63.9 3e-10
CE06958 62.8 7e-10
CE12430 62.8 9e-10
7291055 62.4 1e-09
7294979 62.0 1e-09
SPAC2F7.10 59.3 8e-09
At3g56920 58.9 1e-08
> Hs21450653
Length=337
Score = 130 bits (327), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 81/253 (32%), Positives = 120/253 (47%), Gaps = 40/253 (15%)
Query 60 AIGALHFFVAHAGMAMVLWSFYKTYSTDPGR--------VPDTDEWQRQPPPSL------ 105
A ++ + HA W+++K+ T P + D + ++ + P +
Sbjct 52 AEKVIYLILYHAIFVFFTWTYWKSIFTLPQQPNQKFHLSYTDKERYENEERPEVQKQMLV 111
Query 106 -------ITERKRDGGARYCHKCSKFKPDRCHHSSNSGRCVLKMDHYCPWVANDVGFFNY 158
+ R G R+C +C KPDRCHH S CVLKMDH+CPWV N +GF NY
Sbjct 112 DMAKKLPVYTRTGSGAVRFCDRCHLIKPDRCHHCSVCAMCVLKMDHHCPWVNNCIGFSNY 171
Query 159 KFFFLTLLYSGATLIFVCFTMAATV--------RTSIDDSNISFEQVFFTFLGTALSFFL 210
KFF L YS + C +A TV R + F +F F+ F
Sbjct 172 KFFLQFLAYS----VLYCLYIATTVFSYFIKYWRGELPSVRSKFHVLFLLFVAC---MFF 224
Query 211 LAIVVPFCIFHLVLITSNCTTIE-FCEKRRTGRP--NPYDVGVAENLKQALGDQPLLWPI 267
+++V+ F +H L++ N TT+E FC T P N +++G +N++Q GD+ W I
Sbjct 225 VSLVILFG-YHCWLVSRNKTTLEAFCTPVFTSGPEKNGFNLGFIKNIQQVFGDKKKFWLI 283
Query 268 PVGGPSGDGISFP 280
P+G GDG SFP
Sbjct 284 PIGSSPGDGHSFP 296
> Hs7705949
Length=367
Score = 129 bits (325), Expect = 5e-30, Method: Compositional matrix adjust.
Identities = 87/298 (29%), Positives = 132/298 (44%), Gaps = 49/298 (16%)
Query 14 KRLARFLPMIVICSVMAILYTVYLLYHCLPMFQFEVPPRYRDRGAIAIGALHFFVAHAGM 73
+R+ ++P++ I ++ Y Y + C+ V + + A H
Sbjct 15 RRVLYWIPVVFITLLLGWSYYAYAIQLCI------VSMENTGEQVVCLMAYHLL-----F 63
Query 74 AMVLWSFYKTYSTDPGR--------VPDTDEWQRQPPPSLITE-------------RKRD 112
AM +WS++KT T P + D +R+P E R
Sbjct 64 AMFVWSYWKTIFTLPMNPSKEFHLSYAEKDLLEREPRGEAHQEVLRRAAKDLPIYTRTMS 123
Query 113 GGARYCHKCSKFKPDRCHHSSNSGRCVLKMDHYCPWVANDVGFFNYKFFFLTLLYSGATL 172
G RYC +C KPDRCHH S +C+LKMDH+CPWV N VGF NYKFF L L YS L
Sbjct 124 GAIRYCDRCQLIKPDRCHHCSVCDKCILKMDHHCPWVNNCVGFSNYKFFLLFLAYS---L 180
Query 173 IFVCFTMAATVR-------TSIDDSNISFEQVFFTFLGTALSFFLLAIVVPFCIFHLVLI 225
++ F A ++ + D+ F +F F S L ++ +H L+
Sbjct 181 LYCLFIAATDLQYFIKFWTNGLPDTQAKFHIMFLFFAAAMFSVSLSSLFG----YHCWLV 236
Query 226 TSNCTTIEFCEK---RRTGRPNPYDVGVAENLKQALGDQPLLWPIPVGGPSGDGISFP 280
+ N +T+E R N + +G ++N++Q GD+ W +P+ GDG SFP
Sbjct 237 SKNKSTLEAFRSPVFRHGTDKNGFSLGFSKNMRQVFGDEKKYWLLPIFSSLGDGCSFP 294
> 7303813
Length=338
Score = 128 bits (322), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 91/297 (30%), Positives = 139/297 (46%), Gaps = 50/297 (16%)
Query 18 RFLPMIVICSVMAILYTVYLLYHCLPMFQFEVPPRYRDRGAIAIGALHFFVAHAGMAMVL 77
+++P++ I +V+A Y Y++ C+ R + I L F+ H + + +
Sbjct 21 KWIPVLFITAVIAWSYYAYVVELCI---------RNSENRIGMIFMLLFY--HLFLTLFM 69
Query 78 WSFYKTYSTDPGRVPDTDEWQ----------RQPPPSL-------------ITERKRDGG 114
WS+++T T GR+PD +W+ R P +T R +G
Sbjct 70 WSYWRTIMTSVGRIPD--QWRIPDEEVSRLFRADSPDTQKRILNNFARDLPVTNRTMNGS 127
Query 115 ARYCHKCSKFKPDRCHHSSNSGRCVLKMDHYCPWVANDVGFFNYKFFFLTLLYSGATLIF 174
R+C KC KPDR HH S CVLKMDH+CPWV N V F+NYK+F L L Y+ ++
Sbjct 128 VRFCEKCKIIKPDRAHHCSVCSCCVLKMDHHCPWVNNCVNFYNYKYFVLFLGYALVYCLY 187
Query 175 VCFTMAATVRTSIDDSNISFEQVFFTFLGTALSFFLLAIVVPFCI-------FHLVLITS 227
V FT ++ ++ + Q+ + +G FL I + F I +H+ L+
Sbjct 188 VAFT---SLHDFVEFWKVGAGQLNASGMGRFHILFLFFIAIMFAISLVSLFGYHIYLVLV 244
Query 228 NCTTIEFCEK--RRTGRP--NPYDVGVAENLKQALGDQPLLWPIPVGGPSGDGISFP 280
N TT+E R G P N Y++G N + GD W +PV GDG +FP
Sbjct 245 NRTTLESFRAPIFRVGGPDKNGYNLGRYANFCEVFGDDWQYWFLPVFSSFGDGKTFP 301
> SPBC2F12.15c
Length=329
Score = 119 bits (297), Expect = 9e-27, Method: Compositional matrix adjust.
Identities = 66/213 (30%), Positives = 105/213 (49%), Gaps = 8/213 (3%)
Query 75 MVLWSFYKTYSTDPGRVPDTDEWQRQPPPSLITERKRDGGARYCHKCSKFKPDRCHHSSN 134
M++ + T T PG +T P + ++G +R+C KC ++K DR HH S
Sbjct 59 MIVTCYVLTNLTPPGSPSETS---FDPNSTRQYMTLQNGKSRFCEKCQEYKCDRSHHCSQ 115
Query 135 SGRCVLKMDHYCPWVANDVGFFNYKFFFLTLLYSGATLIFVCFTMAATVRTSIDDSNISF 194
+C+L+MDH+C W N VGF N+KFFFL Y I V ++ + + +
Sbjct 116 CNKCILRMDHHCMWFKNCVGFRNHKFFFLECFYLNLYSICVLYSTFVAITKTFTAEGANI 175
Query 195 EQVFFTFLGTALSFFL-LAIVV-PFCIFHLVLITSNCTTIEFCE---KRRTGRPNPYDVG 249
++ F G +F + ++IV+ F +H L+ N +T+E R T P++VG
Sbjct 176 SAIYLVFWGFLFAFAVGMSIVMTAFTFYHTSLLIHNLSTLESMSSSWSRYTHSTQPFNVG 235
Query 250 VAENLKQALGDQPLLWPIPVGGPSGDGISFPRN 282
EN Q +G P LW +P G+G+ +P N
Sbjct 236 WYENWCQIMGKSPFLWLLPFPNSIGEGVEYPLN 268
> At3g09320
Length=287
Score = 113 bits (283), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 79/225 (35%), Positives = 107/225 (47%), Gaps = 27/225 (12%)
Query 75 MVLWSFYKTYSTDPGRVPDTDEWQRQPPPSLITERKRDGG-ARYCHKCSKFKPDRCHHSS 133
M ++++ DPGRVP + P S + E KR GG RYC KCS FKP R HH
Sbjct 55 MCIYNYSIAVFRDPGRVPLNYMPDVEDPESPVHEIKRKGGDLRYCQKCSHFKPPRAHHCR 114
Query 134 NSGRCVLKMDHYCPWVANDVGFFNYKFFFLTLLYSGATLIFVCFTMAATVRTSIDDSNIS 193
RCVL+MDH+C W+ N VG NYK FF+ ++Y+ ++ + ++ D
Sbjct 115 VCKRCVLRMDHHCIWINNCVGHTNYKVFFVFVVYAVTACVYSLVLLVGSLTVEPQDE--- 171
Query 194 FEQVFFTFLGT--ALSFFLLAIVVPFCI-------FHLVLITSNCTTIE----------F 234
E+ ++L T +S FLL +P I +H+ LI N TTIE
Sbjct 172 -EEEMGSYLRTIYVISAFLL---IPLSIALGVLLGWHIYLILQNKTTIEVYHEGVRAMWL 227
Query 235 CEKRRTGRPNPYDVGVAENLKQALGDQPLLWPIPVGGPSGDGISF 279
EK +PYD+G ENL LG L W P G G+ F
Sbjct 228 AEKGGQVYKHPYDIGAYENLTLILGPNILSWLCPTSRHIGSGVRF 272
> Hs22053668
Length=355
Score = 104 bits (259), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 93/300 (31%), Positives = 139/300 (46%), Gaps = 50/300 (16%)
Query 14 KRLARFLPMIVICSVMAILYTVYLLYHCL-PMFQFEVPPRYRDRGAIAIGALHFFVAHAG 72
+R+ ++P++ I V+ Y Y++ C+ +F E + + + A H F
Sbjct 11 QRVVGWVPVLFITFVVVWSYYAYVVELCVFTIFGNEE----NGKTVVYLVAFHLF----- 61
Query 73 MAMVLWSFYKTYSTDPGRVPD----TDEWQRQPPPSLITERKRD---------------- 112
M +WS++ T T P ++ + + ER+++
Sbjct 62 FVMFVWSYWMTIFTSPASPSKEFYLSNSEKERYEKEFSQERQQEILRRAARALPIYTTSA 121
Query 113 -GGARYCHKCSKFKPDRCHHSSNSGRCVLKMDHYCPWVANDVGFFNYKFFFLTLLYSGAT 171
RYC KC KPDR HH S C+LKMDH+CPWV N VGF NYKFF L LLYS
Sbjct 122 SKTIRYCEKCQLIKPDRAHHCSACDSCILKMDHHCPWVNNCVGFSNYKFFLLFLLYSLLY 181
Query 172 LIFVCFTMAATV--------RTSIDDSNISFEQVFFTFLGTALSFFLLAIVVPFCIFHLV 223
+FV AATV + D+ F V F F +A+ F ++++ F +H
Sbjct 182 CLFV----AATVLEYFIKFWTNELTDTRAKF-HVLFLFFVSAM--FFISVLSLFS-YHCW 233
Query 224 LITSNCTTIE-FCEKRRTGRP--NPYDVGVAENLKQALGDQPLLWPIPVGGPSGDGISFP 280
L+ N TTIE F + P N + +G ++N +Q GD+ W +P+ GDG SFP
Sbjct 234 LVGKNRTTIESFRAPTFSYGPDGNGFSLGCSKNWRQVFGDEKKYWLLPIFSSLGDGCSFP 293
> YNL326c
Length=336
Score = 103 bits (257), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 75/232 (32%), Positives = 108/232 (46%), Gaps = 39/232 (16%)
Query 73 MAMVLWSFYKTYSTDPGR---VPD-------TDEWQRQPPPSLITER----KRDGGARYC 118
+A+ L+++YK + PG PD E + PP +++R K DG R C
Sbjct 47 LAVALYTYYKVIARGPGSPLDFPDLLVHDLKAAENGLELPPEYMSKRCLTLKHDGRFRVC 106
Query 119 HKCSKFKPDRCHHSSNSGRCVLKMDHYCPWVANDVGFFNYKFF----FLTLLYSGATLIF 174
C +KPDRCHH S+ C+LKMDH+CPW A GF N KFF T LY+ LI+
Sbjct 107 QVCHVWKPDRCHHCSSCDVCILKMDHHCPWFAECTGFRNQKFFIQFLMYTTLYAFLVLIY 166
Query 175 VCFTMAATVRTSIDDSNISFEQVFFTFLGTALSFFLLAI-VVPFCIFHLVLITSNCTTIE 233
C+ + T + + + E + F LG AL + I V+ F F + + N TTIE
Sbjct 167 TCYELG----TWFNSGSFNRELIDFHLLGVALLAVAVFISVLAFTCFSIYQVCKNQTTIE 222
Query 234 FCEKRRTGR---------------PNPYDVGVA-ENLKQALGDQPLLWPIPV 269
RR R N +D+G + N + +G L W +P+
Sbjct 223 VHGMRRYRRDLEILNDSYGTNEHLENIFDLGSSMANWQDIMGTSWLEWILPI 274
> At4g22750_2
Length=296
Score = 102 bits (253), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 75/238 (31%), Positives = 104/238 (43%), Gaps = 51/238 (21%)
Query 59 IAIGALHFFVAHAGMAMVLWSFYKTYSTDPGRVP-----DTDEWQRQPPPSLITERK--- 110
+++ L FF H + M+LWS++ TDPG VP + D + + +LI E
Sbjct 52 LSVLVLAFF--HFLLIMLLWSYFSVVVTDPGGVPTGWRPELDIEKSEGNQALIGEASVGD 109
Query 111 -RDGGARYCHKCSKFKPDRCHHSSNSGRCVLKMDHYCPWVANDVGFFNYKFFFLTLLYSG 169
G RYC KC+++KP R HH S GRC+LKMDH+C WV N VG NYK
Sbjct 110 SSSHGVRYCRKCNQYKPPRSHHCSVCGRCILKMDHHCVWVVNCVGANNYK---------- 159
Query 170 ATLIFVCFTMAATVRTSIDDSNISFEQVFFTFLGTALSFFLLAIVVPFCIFHLVLITSNC 229
F + T DS S F L A + +L F I H++L+ N
Sbjct 160 ------SFLLFLNTLTLPSDSKTSLS--CFPVLNIAFALSVLG----FLIMHIMLVARNT 207
Query 230 TTIEFCEKRRTG------------------RPNPYDVGVAENLKQALGDQPLLWPIPV 269
TTIE ++ G PY+VG N +Q G + W +P+
Sbjct 208 TTIEVKQQLSLGFALNLGSLRNLHAYEKHTVNWPYNVGRKTNFEQVFGSDKMYWFVPL 265
> 7302813
Length=338
Score = 94.4 bits (233), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 63/231 (27%), Positives = 93/231 (40%), Gaps = 30/231 (12%)
Query 78 WSFYKTYSTDPGRVPDTDEWQRQPPPSLITERKR-----------------------DGG 114
W++++ P R+PD +W+ P +R DG
Sbjct 66 WTWFRCIFVAPVRIPD--QWKISPEDVDKLKRNDGIEGASRVLNYAARNLPIATCTIDGL 123
Query 115 ARYCHKCSKFKPDRCHHSSNSGRCVLKMDHYCPWVANDVGFFNYKFFFLTLLYSGATLIF 174
RYC C KPDR HH CVLKMDH+CPW+ N V F N+K+F L L Y+ +
Sbjct 124 VRYCKTCWIIKPDRAHHCRTCHMCVLKMDHHCPWIVNCVHFHNFKYFILFLFYAEVYCFY 183
Query 175 VCFTMAATVRTSIDDSNISFE-QVFFTFLGTALSFFLLAIVVPFCIFHLVLITSNCTTIE 233
+ M + + + Q + L + V L+ ++ N TT+E
Sbjct 184 LFCVMVYDLYLICGFEVTALKNQHSWNILQYLVCILFNIFTVIMYTVSLLNVSRNRTTME 243
Query 234 FCEKRR----TGRPNPYDVGVAENLKQALGDQPLLWPIPVGGPSGDGISFP 280
N +++G N + GD+ LWP P+ GDG SFP
Sbjct 244 SAYATYFLLGGKNNNGFNLGYFVNFRDLYGDKWYLWPFPIFSSRGDGFSFP 294
> CE00467_2
Length=290
Score = 89.7 bits (221), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 83/285 (29%), Positives = 119/285 (41%), Gaps = 54/285 (18%)
Query 16 LARFLPMIVICSVMAILYTVYLLYHCLPMFQFEVPPRYRDRGAIAIGALHFFVAHAGMAM 75
L RFLP++++ +A + +Y + L + + P+ ++ F+ +A + +
Sbjct 26 LVRFLPVVLV--SLATGWGIYAYTYELCILSIDNWPQ---------RIIYLFIFYALLIL 74
Query 76 VLWSFYKTYSTDPGRVPDT----------------DEWQRQPPPSLITE--------RKR 111
S+ +T T + P DE Q Q S I R
Sbjct 75 FYTSYLRTVYTKAWKPPQKYCIEGASKATYESVKDDERQLQLFLSDIARERDLTLLVRGF 134
Query 112 DGGARYCHKCSKFKPDRCHHSSNSGRCVLKMDHYCPWVANDVGFFNYKFFFLTL----LY 167
D G R+C KC KPDR HH S +CVLK DH+CPWV N V F NYK+F L L +Y
Sbjct 135 DHGIRFCDKCCCIKPDRSHHCSMCEQCVLKFDHHCPWVNNCVNFGNYKYFILFLALSNIY 194
Query 168 SGATLIFVCFTMAATVRTSIDDSNISFEQVFFTFLGTALSFFLLAIVVPFCIFHLVLITS 227
F CF N F VF FL S L + +HL L
Sbjct 195 ISQAQNFSCFYFYF--------QNGRFPLVFLLFLSCMFSLSLSFLFF----YHLYLTAK 242
Query 228 NCTTIE-FCEKRRTGR--PNPYDVGVAENLKQALGDQPLLWPIPV 269
N TT+E F G+ + ++ G+ N ++ G PL W +PV
Sbjct 243 NRTTVESFRAPMIDGKYAKDAFNHGIRANYREIFGSHPLYWFLPV 287
> CE05134_2
Length=320
Score = 88.6 bits (218), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 56/183 (30%), Positives = 90/183 (49%), Gaps = 13/183 (7%)
Query 105 LITERKRDGGARYCHKCSKFKPDRCHHSSNSGRCVLKMDHYCPWVANDVGFFNYKFFFLT 164
++ E + G +YC++C KPDR H S+ G+C +K DH+CPW+ V NYK+F L
Sbjct 112 VVAECDQVGRLKYCYECGHIKPDRARHCSSCGKCCIKYDHHCPWINMCVTHVNYKYFLLY 171
Query 165 LLYSGATLIFVCFT-MAATVRTSID----DSNISFEQVFFTFL-GTALSFFLLAIVVPFC 218
++Y+ + + T + VR I+ D F F+F+ G ++ L ++
Sbjct 172 IIYTSFLVYWYLLTSLEGAVRYFINQQWTDELGKFLFYLFSFIVGGVFGYYPLGELI--- 228
Query 219 IFHLVLITSNCTTIEFCEKR--RTGRPNPYDVGVAENLKQALGDQPLLWPIPVGGPSGDG 276
IFH LI+ N TT+E + R Y++G N + G LW P+ + DG
Sbjct 229 IFHYQLISLNETTVEQTKPALLRFDNAADYNMGKYNNFQSVFGWG--LWLCPIDSSTQDG 286
Query 277 ISF 279
+ F
Sbjct 287 LHF 289
> At5g04270
Length=284
Score = 88.2 bits (217), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 60/181 (33%), Positives = 87/181 (48%), Gaps = 14/181 (7%)
Query 115 ARYCHKCSKFKPDRCHHSSNSGRCVLKMDHYCPWVANDVGFFNYKFFFLTLLYSGATLIF 174
R C KC +KP R HH RCVLKMDH+C W+ N VG+ NYK FF+ + Y+ I+
Sbjct 104 TRKCDKCFAYKPLRTHHCRVCRRCVLKMDHHCLWINNCVGYANYKAFFILVFYATVASIY 163
Query 175 VCFTMAATVRTSIDD--SNISFEQVFFTFLGTALSFFLLAIVVPFCIFHLVLITSNCTTI 232
+ + D N+ + F G + + + C +H+ LIT N TTI
Sbjct 164 STVLLVCCAFKNGDSYAGNVPL-KTFIVSCGIFMIGLSITLGTLLC-WHIYLITHNMTTI 221
Query 233 EFCEK-------RRTGRP--NPYDVGVAENLKQALGDQPLLWPIPVGGPS-GDGISFPRN 282
E + R++G+ + +DVG +NL LG + W P + DGISF +
Sbjct 222 EHYDSKRASWLARKSGQSYRHQFDVGFYKNLTSVLGPNMIKWLCPTFTRNPEDGISFSAS 281
Query 283 R 283
R
Sbjct 282 R 282
> ECU09g0230
Length=275
Score = 88.2 bits (217), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 56/175 (32%), Positives = 82/175 (46%), Gaps = 12/175 (6%)
Query 116 RYCHKCSKFKPDRCHHSSNSGRCVLKMDHYCPWVANDVGFFNYKFF--FLTLLYSGATLI 173
++C +C+ +KP+R HH S+ G C+ KMDH+C W+ N V + N F FL S LI
Sbjct 82 KFCRRCNNYKPERAHHCSSCGYCIKKMDHHCFWINNCVNYDNQGHFIRFLFFTASANILI 141
Query 174 FVCFTMAATVRTSIDDSNISFEQVFFTFLGTALSFFLLAIVVPFCIFHLVLITSNCTTIE 233
FV ++ + + S + L LS L + F + L SN T IE
Sbjct 142 FVYVSIESAAILLFNHSLEHRRDYYILVLSGMLSLVFLIMTAFFLYLQMRLALSNTTFIE 201
Query 234 FCEKRRTGR-------PNPYDVGVAENLKQALGDQPLLWPIPVGGPSGDGISFPR 281
++ R +PYD G+ NL LG L+ + GP GDGI+F +
Sbjct 202 ELKQESMSRLQGSSILRSPYDRGIMSNLIDTLGPPYTLFLL---GPFGDGITFIK 253
> ECU04g0260
Length=307
Score = 87.4 bits (215), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 59/201 (29%), Positives = 85/201 (42%), Gaps = 32/201 (15%)
Query 109 RKRDGGARYCHKCSKFKPDRCHHSSNSGRCVLKMDHYCPWVANDVGFFNYKFFFLTLLYS 168
R+ + C KC +KP R HH RC LK DH+C + +GF NYKFF+ ++ +
Sbjct 104 RQSIAKVKLCSKCKTYKPPRAHHCGTCKRCYLKYDHHCALLNTCIGFHNYKFFYQFMVLN 163
Query 169 GATLIFVCFTMAATVRTSIDDSNISFEQ--VFFTFLGTALSFFLLAIVVPFCIFHLVLIT 226
+ +F T++ + I S + V + +G F L IFH LI
Sbjct 164 LVSTVFFLVTISIYMMVYIPKSTSHWVNYIVAASLMGIEFIFNL-----SLLIFHTWLIG 218
Query 227 SNCTTIE-----------------FCEKRRTGRP--------NPYDVGVAENLKQALGDQ 261
N TTIE F E T NPY++G +N +Q G
Sbjct 219 MNETTIEHYALNDYISGDHSFSHIFQEGPITTLADSTDRRVLNPYNLGAKQNWRQVFGSD 278
Query 262 PLLWPIPVGGPSGDGISFPRN 282
PL W G+G++FP+N
Sbjct 279 PLDWLTASHSTLGNGMTFPKN 299
> At4g00840
Length=262
Score = 84.3 bits (207), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 69/238 (28%), Positives = 95/238 (39%), Gaps = 56/238 (23%)
Query 75 MVLWSFYKTYSTDPGRVPDTDEWQRQPPPSLITERKRDGGA----RYCHKCSKFKPDRCH 130
M+LWS++ T TDPG VP+ + SL D GA YC KC KP RCH
Sbjct 1 MLLWSYFTTVFTDPGSVPEHFRREMGGGDSLEAGTSTDQGAFGSLGYCTKCRNVKPPRCH 60
Query 131 HSSNSG----------RCVLKMDHYCPWVANDVGFFNYK--------------------- 159
H S RCVLKMDH+C W+ N VG NYK
Sbjct 61 HCSVCKAKSDLFILCQRCVLKMDHHCVWIVNCVGARNYKFFLLFLFYTFLETMLDVIVLL 120
Query 160 -----FFFLTLLYSGATLIFVCFTMAATVRTSIDDSNISFEQVFFTFLG----------- 203
FF + +S + +A S ++V FLG
Sbjct 121 PSFIEFFSQAIKHSSSPGKLASLVLAFGEYKRFFFSFSFSQKVLSFFLGLIFRLYVSYRF 180
Query 204 ----TALSFFLLAIVVPFCIFHLVLITSNCTTIEFCEKRRTGRPNPYDVGVAENLKQA 257
L+F + ++ F + H+ L++SN T++E EK R YD+G +N +Q
Sbjct 181 YPLDAVLNFAFVLSLLCFVVMHISLLSSNTTSVEVHEKNGEVRWK-YDLGKKKNFEQV 237
> YOL003c
Length=378
Score = 82.0 bits (201), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 75/264 (28%), Positives = 105/264 (39%), Gaps = 69/264 (26%)
Query 73 MAMVLWSFYKTYSTDPGR-VPDTDEWQRQPPPSLITERKRDGGARYCHKCSKFKPDRCHH 131
++M+ S+Y T+PGR +P+ +PPP D +C KC +KP+R HH
Sbjct 47 LSMIWLSYYLAICTNPGRPLPNY-----KPPP--------DIWRNFCKKCQSYKPERSHH 93
Query 132 SSNSGRCVLKMDHYCPWVANDVGFFNYKFFFLTLLYSGATLIFVCFTMAATVRTSIDDSN 191
+CVL MDH+CPW N VGF NY F L + T + A + +
Sbjct 94 CKTCNQCVLMMDHHCPWTMNCVGFANYPHFLRFLFWIIVTTSVLFCIQAKRIYFIWQQRH 153
Query 192 I------SFEQVFFTFLGTALSFFLLAIVVPF--CIFHLVL------------------- 224
+ E +F T SF LL I + F C+F+ +L
Sbjct 154 LPGYFFKKSELIFLTISSPLNSFVLLTITILFLRCLFNQILNGRSQIESWDMDRLESLFN 213
Query 225 -------ITSNCTTI-----EFCEKRRT------GRPN-------PYDVGVAENLKQALG 259
+ N I F K+ RP PYD + N LG
Sbjct 214 SGRLTQKLIDNTWRIYPESRSFQNKKDAEEHLTKKRPRFDELVNFPYDFDLYTNALLYLG 273
Query 260 DQPL-LWPIPVGGPSGDGISFPRN 282
P+ LW P G P+GDG +FP+N
Sbjct 274 --PIHLWLWPYGVPTGDGNNFPKN 295
> At3g60800
Length=273
Score = 80.5 bits (197), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 72/244 (29%), Positives = 105/244 (43%), Gaps = 20/244 (8%)
Query 28 VMAILYTVYLLYHCLPMFQFEVPPRYRDRGAIAIGALHFFVA-HAGMAMVLWSFYKTYST 86
++ +L V + Y+ + + + P G ++ AL + H +AM+LWS++ T
Sbjct 20 ILLVLGVVGVTYYAVVLTNYG--PALSQGGLDSLAALTILILFHFLLAMLLWSYFSVVFT 77
Query 87 DPGRVPDTDEWQRQPPPSLITERKRDGGARYCHKCSKFKPDRCHHSSNSGRCVLKMDHYC 146
DPG VP W+ PS ER D S+S +MDH+C
Sbjct 78 DPGVVPP--NWR----PSTDEERGESDPLNSL--------DFVGLQSDSSSSNPRMDHHC 123
Query 147 PWVANDVGFFNYKFFFLTLLYSGATLIFVCFTMAATVRTSIDDSNI--SFEQVFFTFLGT 204
WV N VG NYK+F L L Y+ V + D I + + TFL
Sbjct 124 VWVVNCVGALNYKYFLLFLFYTFLETTLVTLVLMPHFIAFFSDEEIPGTPGTLATTFLAF 183
Query 205 ALSFFLLAIVVPFCIFHLVLITSNCTTIEFCEKRRTGRPNPYDVGVAENLKQALGDQPLL 264
L+ V+ F I H+ L+ N TTIE EK+ T + YD+G +N +Q G
Sbjct 184 VLNLAFALSVMGFLIMHISLVAGNTTTIEAYEKKTTTKWR-YDLGKKKNFEQVFGMDKRY 242
Query 265 WPIP 268
W IP
Sbjct 243 WLIP 246
> At3g56930
Length=477
Score = 78.6 bits (192), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 58/199 (29%), Positives = 86/199 (43%), Gaps = 27/199 (13%)
Query 83 TYSTDPG------RVPDTD----------EWQRQPPPSLITERKRDG-------GARYCH 119
T S DPG R P+TD EW P++ R +D ++C
Sbjct 90 TSSRDPGIVPRSFRPPETDDAPDSTTPSMEWVSGRTPNIRIPRVKDVTVNGHTVKVKFCD 149
Query 120 KCSKFKPDRCHHSSNSGRCVLKMDHYCPWVANDVGFFNYKFFFLTLLYSGATLIFV-CFT 178
C ++P R H S CV + DH+CPWV +G NY+FFF+ + S I+V F+
Sbjct 150 TCLLYRPPRASHCSICNNCVQRFDHHCPWVGQCIGVRNYRFFFMFISTSTTLCIYVFAFS 209
Query 179 MAATVRTSIDDSNISFEQVFFTFLGTAL---SFFLLAIVVPFCIFHLVLITSNCTTIEFC 235
+ +D+ ++ + L L F + V IFH LI +N TT E
Sbjct 210 WLNIFQRHMDEKISIWKAISKDVLSDILIVYCFITVWFVGGLTIFHSYLICTNQTTYENF 269
Query 236 EKRRTGRPNPYDVGVAENL 254
R + NPY+ G+ N+
Sbjct 270 RYRYDKKENPYNKGILGNI 288
> At3g48760
Length=470
Score = 78.6 bits (192), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 47/150 (31%), Positives = 74/150 (49%), Gaps = 12/150 (8%)
Query 116 RYCHKCSKFKPDRCHHSSNSGRCVLKMDHYCPWVANDVGFFNYKFFFLTLLYSGATLIFV 175
+YC C ++P R H S CV K DH+CPW+ +G NY+F+F+ +L S I+V
Sbjct 152 KYCDTCMLYRPPRASHCSICNNCVEKFDHHCPWLGQCIGLRNYRFYFMFVLCSTLLCIYV 211
Query 176 CFTMAATVRTSIDDSNISFEQVFFTFLGTALSFFLLAIVVPFCI--------FHLVLITS 227
V+ +D NI+ ++ +FL T S L+ I C+ FHL L+++
Sbjct 212 HVFCWIYVKRIMDSENIN---IWKSFLKTPASIALI-IYTFICVWFVGGLTCFHLYLMST 267
Query 228 NCTTIEFCEKRRTGRPNPYDVGVAENLKQA 257
N +T E R NP++ G+ N +
Sbjct 268 NQSTYENFRYRYDRHENPFNKGIVGNFMEV 297
> At5g41060
Length=410
Score = 77.4 bits (189), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 65/258 (25%), Positives = 104/258 (40%), Gaps = 35/258 (13%)
Query 27 SVMAILYTVYLLYHCLPMFQFEVPPRYRDRGAIAIGALHFFVAHAGMAMVLWSFYKTYST 86
V ++ T+ L+ + +F V + D + + G VA L T
Sbjct 41 DVRSLGLTISLIVAPVTIFCIFVASKLMDDFSDSWGVSIILVAVVFTIYDLILLMLTSGR 100
Query 87 DPGRVP-----------------DTDEWQRQPPPSLITERKRDGGARYCHKCSKFKPDRC 129
DPG +P T + R P + + +YC C ++P RC
Sbjct 101 DPGIIPRNSHPPEPEVVDGNTGSGTSQTPRLPRVKEVEVNGKVFKVKYCDTCMLYRPPRC 160
Query 130 HHSSNSGRCVLKMDHYCPWVANDVGFFNYKFFFLTLLYSGATLIFV---CFTMAATVRTS 186
H S CV + DH+CPWV + NY+FFF+ + + ++V C ++ S
Sbjct 161 SHCSICNNCVERFDHHCPWVGQCIAQRNYRFFFMFVFSTTLLCVYVFAFCCVYIKKIKES 220
Query 187 IDDSNI--------SFEQVFFTFLGTALSFFLLAIVVPFCIFHLVLITSNCTTIEFCEKR 238
D S + S + +TF+ T FF+ + FHL LI++N TT E
Sbjct 221 EDISILKAMLKTPASIALILYTFIST---FFVGGLTC----FHLYLISTNQTTYENFRYS 273
Query 239 RTGRPNPYDVGVAENLKQ 256
NP++ GV +N K+
Sbjct 274 YDRHSNPHNKGVVDNFKE 291
> Hs14150106
Length=296
Score = 76.6 bits (187), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 67/193 (34%), Positives = 92/193 (47%), Gaps = 35/193 (18%)
Query 118 CHKCSKFKPDRCHHSSNSGRCVLKMDHYCPWVANDVGFFNYKFFF-------LTLLYS-- 168
C KC KP R HH S RCVLKMDH+CPW+ N VG +N+++FF L +Y
Sbjct 92 CKKCIYPKPARTHHCSICNRCVLKMDHHCPWLNNCVGHYNHRYFFSFCFFMTLGCVYCSY 151
Query 169 GATLIFVCFTMAATVRTSIDDSNISFEQ-------VFFTFLGTALSFFLLAIVVPFCIFH 221
G+ +F A SF + V+ FL ++++ L A+ V +H
Sbjct 152 GSWDLFREAYAAIETYHQTPPPTFSFRERMTHKSLVYLWFLCSSVALALGALTV----WH 207
Query 222 LVLITSNCTTIEFC----EKRR---TGR--PNPYDVGVAENLKQALG-DQPLLWPIPVGG 271
VLI+ T+IE E+RR GR NPY+ G +N K LG D W V
Sbjct 208 AVLISRGETSIERHINKKERRRLQAKGRVFRNPYNYGCLDNWKVFLGVDTGRHWLTRVLL 267
Query 272 PS-----GDGISF 279
PS G+G+S+
Sbjct 268 PSSHLPHGNGMSW 280
> Hs14761406
Length=382
Score = 76.3 bits (186), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 65/211 (30%), Positives = 92/211 (43%), Gaps = 32/211 (15%)
Query 77 LWSFYKTYSTDPGRVPDT--DEW---------------QRQPPPSLITERKRDGGA---R 116
+ + +T +DPG +P DE Q Q PP I + + +
Sbjct 98 MATLLRTSFSDPGVIPRALPDEAAFIEMEIEATNGAVPQGQRPPPRIKNFQINNQIVKLK 157
Query 117 YCHKCSKFKPDRCHHSSNSGRCVLKMDHYCPWVANDVGFFNYKFFFLTLLYSGATLIFV- 175
YC+ C F+P R H S CV + DH+CPWV N VG NY++F+L +L I+V
Sbjct 158 YCYTCKIFRPPRASHCSICDNCVERFDHHCPWVGNCVGKRNYRYFYLFILSLSLLTIYVF 217
Query 176 CFTMAATVRTSIDDSNISFEQVFFTFLGTALS----FFLLAIVVPFCIFHLVLITSNCTT 231
F + V ++ I F + GT L FF L VV FH L+ N TT
Sbjct 218 AFNI---VYVALKSLKIGFLETLKETPGTVLEVLICFFTLWSVVGLTGFHTFLVALNQTT 274
Query 232 IEFCEKRRTGR---PNPYDVG-VAENLKQAL 258
E + TG+ NPY G + +N + L
Sbjct 275 NEDIKGSWTGKNRVQNPYSHGNIVKNCCEVL 305
> At5g50020
Length=414
Score = 75.9 bits (185), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 45/145 (31%), Positives = 69/145 (47%), Gaps = 6/145 (4%)
Query 115 ARYCHKCSKFKPDRCHHSSNSGRCVLKMDHYCPWVANDVGFFNYKFFFLTLLYSGATLIF 174
+YC C ++P RC H S CV + DH+CPWV +G NY++FF+ + S L
Sbjct 135 VKYCDTCMLYRPPRCSHCSICNNCVERFDHHCPWVGQCIGVRNYRYFFM-FVSSATILCI 193
Query 175 VCFTMAAT-VRTSIDDSNISFEQVF----FTFLGTALSFFLLAIVVPFCIFHLVLITSNC 229
F+M+A ++ +D+ + + + + F L V FHL LI++N
Sbjct 194 YIFSMSALYIKVLMDNHQGTVWRAMRESPWAVMLMIYCFISLWFVGGLTGFHLYLISTNQ 253
Query 230 TTIEFCEKRRTGRPNPYDVGVAENL 254
TT E R R N Y+ G + N
Sbjct 254 TTYENFRYRSDNRINVYNRGCSNNF 278
> Hs7705801
Length=382
Score = 75.9 bits (185), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 61/181 (33%), Positives = 85/181 (46%), Gaps = 18/181 (9%)
Query 89 GRVPDTDEWQRQPPPSLITERKRDG--GARYCHKCSKFKPDRCHHSSNSGRCVLKMDHYC 146
G VP +QR PPP + + + +YC+ C F+P R H S CV + DH+C
Sbjct 132 GAVPG---YQR-PPPRIKNFQINNQIVKLKYCYTCKIFRPPRASHCSICDNCVERFDHHC 187
Query 147 PWVANDVGFFNYKFFFLTLLYSGATLIFV-CFTMAATVRTSIDDSNISFEQVFFTFLGTA 205
PWV N VG NY++F+L +L I+V F + V ++ I F + GT
Sbjct 188 PWVGNCVGKRNYRYFYLFILSLSLLTIYVFAFNI---VYVALKSLKIGFLETLKETPGTV 244
Query 206 LS----FFLLAIVVPFCIFHLVLITSNCTTIEFCEKRRTGR---PNPYDVG-VAENLKQA 257
L FF L VV FH L+ N TT E + TG+ NPY G + +N +
Sbjct 245 LEVLICFFTLWSVVGLTGFHTFLVALNQTTNEDIKGSWTGKNRVQNPYSHGNIVKNCCEV 304
Query 258 L 258
L
Sbjct 305 L 305
> Hs8923255
Length=308
Score = 75.5 bits (184), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 65/207 (31%), Positives = 86/207 (41%), Gaps = 17/207 (8%)
Query 74 AMVLWSFYKTYSTDPGRVPDTDEWQRQPPPSLITERKRDGGARY-CHKCSKFKPDRCHHS 132
+ L S +T TDPG VP + + + + + + G Y C KC KP+R HH
Sbjct 91 VLALSSHLRTMLTDPGAVPKGNATKEY----MESLQLKPGEVIYKCPKCCCIKPERAHHC 146
Query 133 SNSGRCVLKMDHYCPWVANDVGFFNYKFFFLTLLY----SGATLIFVCFTMAATVR---T 185
S RC+ KMDH+CPWV N VG N +FF L +Y S LI F + VR T
Sbjct 147 SICKRCIRKMDHHCPWVNNCVGEKNQRFFVLFTMYIALSSVHALILCGFQFISCVRGQWT 206
Query 186 SIDDSNISFEQVFFTFLGTALSFFLLAIVVPFCIFHLVLITSNCTTIEFCEKRRTGRPNP 245
D + + FL F V F I S C E+ ++ +P
Sbjct 207 ECSDFSPPITVILLIFLCLEGLLFFTFTAVMFG----TQIHSICNDETEIERLKSEKPTW 262
Query 246 YDVGVAENLKQALGDQP-LLWPIPVGG 271
E +K G P LLW P G
Sbjct 263 ERRLRWEGMKSVFGGPPSLLWMNPFVG 289
> CE21719
Length=393
Score = 75.5 bits (184), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 73/264 (27%), Positives = 111/264 (42%), Gaps = 27/264 (10%)
Query 14 KRLARFLP------MIVICSVMAILYTVYLLYHCLPMFQFEVPPRYRDRGAIAIGALHFF 67
KR++ LP +I+ CSV + +++ Q+ P IAI + F
Sbjct 3 KRISALLPAAIAWALIIGCSVSFFWFIAPQIWN-----QWHTPGLI----LIAIDVVLFL 53
Query 68 VAHAG--MAMVLWSFYKTY---STDPGRVPDTDEWQRQPPPSLITERKRDGGARYCHKCS 122
+ + MAM+L Y S +P +V D R P + ++C C
Sbjct 54 MVSSNLLMAMLLDPAVHPYAIGSEEPTQVDDL----RAPLYKNVDINGITVRMKWCVTCK 109
Query 123 KFKPDRCHHSSNSGRCVLKMDHYCPWVANDVGFFNYKFFFLTLLYSGATLIFVCFTMAAT 182
++P R H S RC+ DH+CPWV N VG NY++FF L +++V F A
Sbjct 110 FYRPPRSSHCSVCNRCIETFDHHCPWVHNCVGKRNYRYFFFFLCSLSIHMMYVFFLCFAY 169
Query 183 VRTSIDDS---NISFEQVFFTFLGTALSFFLLAIVVPFCIFHLVLITSNCTTIEFCEKRR 239
V + D + +I + AL L V+ +FHLVL+ TT E +
Sbjct 170 VWSGSDTNARDHILSPPYLCAIVLLALCAVLCVPVIGLTVFHLVLVARGRTTNEQVTGKF 229
Query 240 TGRPNPYDVGVAENLKQALGDQPL 263
T NP+ +G N K+ L L
Sbjct 230 TSGYNPFTIGCWGNCKKTLCHSQL 253
> Hs22041784
Length=396
Score = 75.5 bits (184), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 55/192 (28%), Positives = 81/192 (42%), Gaps = 26/192 (13%)
Query 61 IGALHFFVAHAGMAMVLWSFYKTYSTDPGRVP---------------DTDEWQRQPPPSL 105
I A+ FF V+ +T TDPG +P +T +PPP
Sbjct 127 IAAILFF-------FVMSCLLQTSFTDPGILPRATVCEAAALEKQIDNTGSSTYRPPPRT 179
Query 106 --ITERKRDGGARYCHKCSKFKPDRCHHSSNSGRCVLKMDHYCPWVANDVGFFNYKFFFL 163
+ + +YC C F+P R H S CV + DH+CPWV N VG NY+FF+
Sbjct 180 REVLINGQMVKLKYCFTCKMFRPPRTSHCSVCDNCVERFDHHCPWVGNCVGRRNYRFFYA 239
Query 164 TLLYSG--ATLIFVCFTMAATVRTSIDDSNISFEQVFFTFLGTALSFFLLAIVVPFCIFH 221
+L IF C T+R + + ++ + L + FF + ++ FH
Sbjct 240 FILSLSFLTAFIFACVVTHLTLRAQGSNFLSTLKETPASVLELVICFFSIWSILGLSGFH 299
Query 222 LVLITSNCTTIE 233
L+ SN TT E
Sbjct 300 TYLVASNLTTNE 311
> 7299620
Length=427
Score = 73.9 bits (180), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 60/220 (27%), Positives = 87/220 (39%), Gaps = 42/220 (19%)
Query 86 TDPGRVPDTDEWQRQPPPSLITERKRDGGARYCHKCSKFKPDRCHHSSNSGRCVLKMDHY 145
T PG +P +W + P K +YC KC +K R HH RCV KMDH+
Sbjct 70 TGPGLMPK--QWHPKDP-------KDAQFLQYCKKCEGYKAPRSHHCRKCDRCVKKMDHH 120
Query 146 CPWVANDVGFFNYKFFFLTLLYS-----GATLIFVCFTMAATVR--------TSIDDSNI 192
CPW+ + VG+ N+ +F LL+S T++ C R +
Sbjct 121 CPWINHCVGWANHAYFTYFLLFSILGSLQGTVVLCCSFWRGIYRYYYLTHGLAHLASVQF 180
Query 193 SFEQVFFTFLGTALSFFLLAIVVPFCIFHLVLITSNCTTIE--FCEK---RRTGRPN--- 244
+ + LG L+ ++ + L I +N T IE EK RR +
Sbjct 181 TLLSIIMCILGMGLAIGVVIGLSMLLFIQLKTIVNNQTGIEIWIVEKAIYRRYRNADCDD 240
Query 245 ----PYDVGVAENLKQALGDQPLLWPIPVGGPSGDGISFP 280
PYD+G NL+ D+ GDGI +P
Sbjct 241 EFLYPYDLGWRANLRLVFNDE--------CQKRGDGIEWP 272
> Hs11968053
Length=413
Score = 73.6 bits (179), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 80/319 (25%), Positives = 125/319 (39%), Gaps = 74/319 (23%)
Query 2 SLTMMERREEAGKRLARFLPMIV-----ICSVMAILYTVYLLYHCLPMFQFEVPPRYRDR 56
S+ E +E KRL + P+I ICS MA++ +V + P +
Sbjct 6 SVIKFENLQEL-KRLCHWGPIIALGVIAICSTMAMIDSVLWYW-----------PLHTTG 53
Query 57 GAIAIGALHFFVAHAGMAMVLWSFYKTYSTDPGRVPDTDEWQRQPPPSLITERKRDGGAR 116
G++ +F + M+L++++ PG VP W+ + + +
Sbjct 54 GSV-----NFIMLINWTVMILYNYFNAMFVGPGFVPLG--WKPEISQDTMY-------LQ 99
Query 117 YCHKCSKFKPDRCHHSSNSGRCVLKMDHYCPWVANDVGFFNYKFFFLTLLYSGATLIFVC 176
YC C +K R HH RCV+KMDH+CPW+ N G+ N+ F L LL + I
Sbjct 100 YCKVCQAYKAPRSHHCRKCNRCVMKMDHHCPWINNCCGYQNHASFTLFLLLAPLGCIHAA 159
Query 177 FTMAATV------RTSIDDSNISFEQ----------VFFTFLGTALSFFLLAIVVPFCI- 219
F T+ R S + + + V F A + F L + + I
Sbjct 160 FIFVMTMYTQLYHRLSFGWNTVKIDMSAARRDPLPIVPFGLAAFATTLFALGLALGTTIA 219
Query 220 ------FHLVLITSNCTTIE-FCEKRRTGRPN----------PYDVGVA-ENLKQALGDQ 261
+ +I N T+IE + E++ R PYD+G N KQ
Sbjct 220 VGMLFFIQMKIILRNKTSIESWIEEKAKDRIQYYQLDEVFVFPYDMGSRWRNFKQV---- 275
Query 262 PLLWPIPVGGPSGDGISFP 280
W G P GDG+ +P
Sbjct 276 -FTWS---GVPEGDGLEWP 290
> CE28292
Length=260
Score = 73.6 bits (179), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 41/107 (38%), Positives = 53/107 (49%), Gaps = 8/107 (7%)
Query 64 LHFFVAHAGMAMVLWSFYKTYSTDPGRVPDTDEWQRQPPPSLITERKRDGGARY---CHK 120
+F V + + + S KT +TDPG V D I + G + C K
Sbjct 34 FNFIVFESFSVLAVISHLKTMTTDPGAVAKGD-----CTDETIERMQLINGQQTIYKCQK 88
Query 121 CSKFKPDRCHHSSNSGRCVLKMDHYCPWVANDVGFFNYKFFFLTLLY 167
C+ KPDR HH S RC+ +MDH+CPWV N VG N KFF L +Y
Sbjct 89 CASIKPDRAHHCSVCERCIRRMDHHCPWVNNCVGEGNQKFFVLFTMY 135
> 7303015
Length=293
Score = 73.2 bits (178), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 69/262 (26%), Positives = 105/262 (40%), Gaps = 31/262 (11%)
Query 24 VICSVMAILYTVYLLYHCLPMFQFEVPPRYRDRGAIAIGALHFFVAHAGMAMVLWSFYKT 83
++C +M L ++ + + M +P Y ++ + A + S +T
Sbjct 39 IVCVIMTWLLILFAEF--VVMRLILLPSNY-----TVFSTINMIIFQALAFLAFASHIRT 91
Query 84 YSTDPGRVPDTDEWQRQPPPSLITERK-RDGGARY-CHKCSKFKPDRCHHSSNSGRCVLK 141
+DPG VP + +I + R+G Y C KC KP+R HH S RC+ K
Sbjct 92 MLSDPGAVPRGN-----ATKEMIEQMGYREGQMFYKCPKCCSIKPERAHHCSVCQRCIRK 146
Query 142 MDHYCPWVANDVGFFNYKFFFLTLLY----SGATLIFVCFTMAATVRTSIDDSN------ 191
MDH+CPWV N VG N K+F L Y S TL V A V+ +
Sbjct 147 MDHHCPWVNNCVGENNQKYFVLFTFYIASISVHTLFLVLTQFAECVKNDWRTCSPYSPPA 206
Query 192 ISFEQVFFTFLGTALSFFLLAIVVPFCIFHLVLITSNCTTIEFCEKRRTGRPNPYDVGVA 251
F +F TF G F + ++ L I ++ T IE +K +
Sbjct 207 TIFLLLFLTFEGLMFGIFTIIMLAT----QLTAILNDQTGIEQLKKEEARWAKKSRL--- 259
Query 252 ENLKQALGDQPLLWPIPVGGPS 273
++++ G L W P PS
Sbjct 260 KSIQSVFGRFSLAWFSPFTEPS 281
> CE19697
Length=302
Score = 72.8 bits (177), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 76/296 (25%), Positives = 112/296 (37%), Gaps = 54/296 (18%)
Query 18 RFLPMIVICSVMAILYTVY-----LLYHCLPMFQFEVPPRYRDRGAIAIGALHFFVAHAG 72
++ P ++C M L Y L++ LP F G H V +
Sbjct 3 KWDPCGLVCVCMIYLLIAYADYVILIWLLLPTF-----------GHSIWTVFHGAVFNCL 51
Query 73 MAMVLWSFYKTYSTDPGRVP-DTDEWQRQPPPSLIT-----------------ERKRDGG 114
+A + + + +DPG VP + + Q P P + R
Sbjct 52 LATTIVAHTRAMLSDPGTVPISSSKGQNTPNPVFSSDEEDESDEEAVFRHDHLNRSSATE 111
Query 115 ARYCHKCSKFKPDRCHHSSNSGRCVLKMDHYCPWVANDVGFFNYKFFFLTLLYSGA---- 170
C +C +P R HH RCV KMDH+CPWV N VG +N K+F + Y GA
Sbjct 112 WTMCTRCDSLRPPRAHHCRVCKRCVRKMDHHCPWVNNCVGEYNQKWFLQFIFYVGASSAY 171
Query 171 TLIFVCFTMA---ATVRTSID---DSNISFEQVFFTFLGTALSFFLLAIVVPFCIFHLVL 224
+L+ +C A T I N+ +V + + S V+ L
Sbjct 172 SLLVLCLCWVWHDAYGMTGIKGPLGENLYHAKVIHSIMLAMESALFGLFVLAVSCDQLGA 231
Query 225 ITSNCTTIEFCEKR------RTGRPNPYDVGVAENLKQALGDQP-LLWPIPVGGPS 273
I ++ T IE ++R + RP V + +KQ G P LLW IP P
Sbjct 232 IFTDETAIESVQRRGRNYLASSRRPRNSKVAM---MKQVCGPGPKLLWLIPCANPQ 284
> CE20263
Length=368
Score = 72.4 bits (176), Expect = 9e-13, Method: Compositional matrix adjust.
Identities = 64/228 (28%), Positives = 96/228 (42%), Gaps = 32/228 (14%)
Query 30 AILYTVYLLYHCLPM-FQFEVPPRYRDRGAIAIGALHFFVAHAGMAMVLWSFYKTYSTDP 88
A + TV L+ L + F F+ P + AI I VA +V+ +F+ T TDP
Sbjct 64 AFVVTVILMIATLTVYFVFDAPFLWGYSPAIPI------VAAVLSLIVITNFFATSFTDP 117
Query 89 GRVPDTD-----EWQRQ--------------PPPSLITERKRDGGARYCHKCSKFKPDRC 129
G +P D E RQ P + +YC C ++P RC
Sbjct 118 GILPRVDNIEIIEMDRQQANGNGINDVAHLRPRFQDVVVNGEHVKMKYCTTCRLYRPPRC 177
Query 130 HHSSNSGRCVLKMDHYCPWVANDVGFFNYKFFFLTLLYSGATLIFVCFTMAATVRTSIDD 189
H + CVL DH+CPWV N +G NY +F+ ++ + L+ F A T S+
Sbjct 178 SHCAICDNCVLMFDHHCPWVGNCIGLRNYTYFY-RFVFCLSILVIYLFASAVT-HISLLA 235
Query 190 SNISFEQVFFTFLGTA----LSFFLLAIVVPFCIFHLVLITSNCTTIE 233
+ F V G+A + FF ++ FH L+ ++ TT E
Sbjct 236 QEMPFGDVMRKTPGSAVVIVICFFTTWSIIGLACFHTYLLCADLTTNE 283
> CE20517
Length=240
Score = 72.4 bits (176), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 54/173 (31%), Positives = 75/173 (43%), Gaps = 20/173 (11%)
Query 118 CHKCSKFKPDRCHHSSNSGRCVLKMDHYCPWVANDVGFFNYKFFFLTLLYSGA----TLI 173
C +C +P R HH RCV KMDH+CPWV N VG +N K+F + Y GA +L+
Sbjct 53 CTRCDSLRPPRAHHCRVCKRCVRKMDHHCPWVNNCVGEYNQKWFLQFIFYVGASSAYSLL 112
Query 174 FVCFTMA---ATVRTSID---DSNISFEQVFFTFLGTALSFFLLAIVVPFCIFHLVLITS 227
+C A T I N+ +V + + S V+ L I +
Sbjct 113 VLCLCWVWHDAYGMTGIKGPLGENLYHAKVIHSIMLAMESALFGLFVLAVSCDQLGAIFT 172
Query 228 NCTTIEFCEKR------RTGRPNPYDVGVAENLKQALGDQP-LLWPIPVGGPS 273
+ T IE ++R + RP V + +KQ G P LLW IP P
Sbjct 173 DETAIESVQRRGRNYLASSRRPRNSKVAM---MKQVCGPGPKLLWLIPCANPQ 222
> Hs20482198
Length=715
Score = 71.2 bits (173), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 47/147 (31%), Positives = 69/147 (46%), Gaps = 2/147 (1%)
Query 116 RYCHKCSKFKPDRCHHSSNSGRCVLKMDHYCPWVANDVGFFNYKFFFLTLLYSGATLIFV 175
++C C ++P RC H S CV + DH+CPWV N +G NY++FFL LL A ++ V
Sbjct 104 KWCATCRFYRPPRCSHCSVCDNCVEEFDHHCPWVNNCIGRRNYRYFFLFLLSLTAHIMGV 163
Query 176 CFTMAATVRTSIDDSNISFEQVFFTFLGTALSFFLLAIVVPFCIFHLVLITSNCTTIEFC 235
V I++ + V + A FF+ V FH+VL+ TT E
Sbjct 164 FGFGLLYVLYHIEELSGVRTAVTMAVMCVAGLFFI--PVAGLTGFHVVLVARGRTTNEQV 221
Query 236 EKRRTGRPNPYDVGVAENLKQALGDQP 262
+ G NP+ G N+ + L P
Sbjct 222 TGKFRGGVNPFTNGCCNNVSRVLCSSP 248
> Hs22062547
Length=284
Score = 70.5 bits (171), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 56/196 (28%), Positives = 86/196 (43%), Gaps = 26/196 (13%)
Query 105 LITERKRDGGARYCHKCSKFKPDRCHHSSNSGRCVLKMDHYCPWVANDVGFFNYKFFFLT 164
++ R G YC++C P R H S C+L+ DH+C + VGF NY+ F
Sbjct 83 MLAGRGLGQGWAYCYQCQSQVPPRSGHCSACRVCILRRDHHCRLLGRCVGFGNYRPFLCL 142
Query 165 LLYSGATLIFVCF----TMAATVR--TSIDDSNISFEQVFFTFLG-TALSFFLLAIVVPF 217
LL++ L+ V ++A +R T + + + G +L+ F LA V
Sbjct 143 LLHAAGVLLHVSVLLGPALSALLRAHTPLHMAALLLLPWLMLLTGRVSLAQFALAFVTDT 202
Query 218 CI-----------FHLVLITSNCTTIEFCEKRRTGRPNPYDVGVAENLKQALGDQ---PL 263
C+ FH +L+ TT E+ + + YD+G NL+ ALG +
Sbjct 203 CVAGALLCGAGLLFHGMLLLRGQTTWEWARGQHS-----YDLGPCHNLQAALGPRWALVW 257
Query 264 LWPIPVGGPSGDGISF 279
LWP GDGI+F
Sbjct 258 LWPFLASPLPGDGITF 273
> At4g24630
Length=374
Score = 69.7 bits (169), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 62/237 (26%), Positives = 101/237 (42%), Gaps = 36/237 (15%)
Query 17 ARFLPMIVICSVM-AILYTVYLLYHCLPMFQFEVPPRYRDRGAIAIGALHFFVAHAGMAM 75
AR LP+ ++ ++ +L+ V++ H + E P Y AI + A+ F +
Sbjct 27 ARSLPLTLLLIIVPVVLFCVFVARH----LRHEFSP-YNAGYAIMVVAILFTI------Y 75
Query 76 VLWSFYKTYSTDPGRVPDTDE--------------WQRQPPPSLITERKR---DGGA--- 115
VL + T + DPG VP RQ P I K +G +
Sbjct 76 VLILLFFTSARDPGIVPRNSHPPEEDLRYETTVSADGRQTPSVQIPRTKEVIVNGVSVRV 135
Query 116 RYCHKCSKFKPDRCHHSSNSGRCVLKMDHYCPWVANDVGFFNYKFFFLTLLYSGATLIFV 175
+YC C ++P RC H S CV + DH+CPWV +G NY++FF+ + S I++
Sbjct 136 KYCDTCMLYRPPRCSHCSICNNCVERFDHHCPWVGQCIGLRNYRYFFMFVSSSTLLCIYI 195
Query 176 CFTMAATVRTSIDDSNIS----FEQVFFTFLGTALSFFLLAIVVPFCIFHLVLITSN 228
A ++ +D + ++ + + F L V FHL LI++N
Sbjct 196 FSMSAVYIKILMDHQQATVWRAMKESPWAVVLMIYCFIALWFVGGLTAFHLYLISTN 252
> Hs7706133
Length=327
Score = 69.3 bits (168), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 40/94 (42%), Positives = 51/94 (54%), Gaps = 5/94 (5%)
Query 75 MVLWSFYKTYSTDPGRVPDTDEWQRQPPPSLITERKRDGGARY-CHKCSKFKPDRCHHSS 133
+ L S + TDPG VP + ++ SL + + G Y C KC KPDR HH S
Sbjct 89 LALASHCRAMLTDPGAVPKGNA-TKEFIESL---QLKPGQVVYKCPKCCSIKPDRAHHCS 144
Query 134 NSGRCVLKMDHYCPWVANDVGFFNYKFFFLTLLY 167
RC+ KMDH+CPWV N VG N K+F L +Y
Sbjct 145 VCKRCIRKMDHHCPWVNNCVGENNQKYFVLFTMY 178
> 7297775
Length=276
Score = 68.6 bits (166), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 56/211 (26%), Positives = 83/211 (39%), Gaps = 34/211 (16%)
Query 79 SFYKTYSTDPGRVP---------DTDEWQRQPPPSLITERKRDGGARYCHKCSKFKPDRC 129
S K +DPG VP D + PP C +C ++P R
Sbjct 59 SHSKAVFSDPGTVPLPANRLDFSDLHTTNKNNPPP---GNGHSSEWTVCTRCETYRPPRA 115
Query 130 HHSSNSGRCVLKMDHYCPWVANDVGFFNYKFFFLTLLYSGATLIFVCFTMAATVRTSIDD 189
HH RC+ +MDH+CPW+ N VG N K+F L+Y + +++A V + +
Sbjct 116 HHCRICKRCIRRMDHHCPWINNCVGERNQKYFLQFLIYVA---LLSLYSIALIVGSWVWP 172
Query 190 SNISFEQVFFTFLGTALSFFLLAIVVPFCIF-------HLVLITSNCTTIEFCEKRRTGR 242
+ V T L S L+ + F +F L I + T +E +++ T R
Sbjct 173 CEECSQNVIETQLRMIHSVILMLVSALFGLFVTAIMVDQLHAILYDETAVEAIQQKGTYR 232
Query 243 PNPYDVGVAENLKQALGD-----QPLLWPIP 268
PN Q L D P LW +P
Sbjct 233 PN-------RRKYQLLADVFGRGHPALWLLP 256
> SPBC17A3.11
Length=274
Score = 68.2 bits (165), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 70/280 (25%), Positives = 118/280 (42%), Gaps = 30/280 (10%)
Query 5 MMERREEAGKRLARFLPMIVICSVMAIL----YTVYLLYHCLPM-FQFEVPPRYRDRGAI 59
M EE +R ++ + + AIL YTV++ + + ++ YR+ G
Sbjct 1 MSTNLEEKQRRFSKLQKTLGVVFPAAILLSTGYTVWVFIALICVDSNIKIRNGYRNLGGG 60
Query 60 AIGALHFFVAHAGMAMVLWSFYKTYSTDPGRVPDT--DEWQRQPPPSLITERKRDGGARY 117
+ + FF+ +G+A +S+++ + P +T + P L +G R
Sbjct 61 IVLIIFFFIT-SGLAY--FSYFRVLFSSPSFCGNTLYTYYGFDNPIFLCGP---NGAPRM 114
Query 118 CHKCSKFKPDRCHHSSNSGRCVLKMDHYCPWVANDVGFFNYKFFFLTLLYSGATLIFVCF 177
C C + PDR HHS S RC+ K DHYC +V DV F N KFF+ L Y + V
Sbjct 115 CGTCKCWLPDRSHHSRVSMRCIRKFDHYCSFVGKDVCFSNQKFFYQFLFYGFSAACMVLI 174
Query 178 TMAATVRTSIDDSNISFEQVFFTFLGTALSFFLLAIVVPFCIFHLVLITSN-----CTTI 232
+ A + + ++ +F FL ++V + L+ I S+ T I
Sbjct 175 STAIMISRTYHYRSLPGTWIFVLVFSAFGVLFLGVMLVRHTGYLLLNINSHEAKNWKTRI 234
Query 233 --------EFCEKRRTGRPN----PYDVGVAENLKQALGD 260
E + R + + P+D G +EN + +GD
Sbjct 235 YSFSVFFPEHMDSRVLVQSDPGDLPWDRGYSENWRAVMGD 274
> 7301549
Length=357
Score = 66.6 bits (161), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 28/58 (48%), Positives = 39/58 (67%), Gaps = 0/58 (0%)
Query 118 CHKCSKFKPDRCHHSSNSGRCVLKMDHYCPWVANDVGFFNYKFFFLTLLYSGATLIFV 175
C KC KP R HH S RC+LKMDH+CPW+ N VG+ N+++FFL + Y+ +F+
Sbjct 115 CGKCIAPKPPRTHHCSICNRCILKMDHHCPWLNNCVGYGNHRYFFLYMTYTTLGCLFL 172
> 7294202
Length=637
Score = 65.9 bits (159), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 67/265 (25%), Positives = 106/265 (40%), Gaps = 49/265 (18%)
Query 14 KRLARFLPMIVICSVMAILYTVYLLYHCLPMFQFEVPPRYRDRGAIAIGALHFFVAHAGM 73
+ L LP+ V + A Y +L+Y D + F ++
Sbjct 344 EHLMALLPLSVYLATKAWFYVTWLMY--------------IDDAVSFTATVCFLIS---- 385
Query 74 AMVLW-SFYKTYSTDPGRVPDTDEWQRQPPPSLITERKRDG--GARYCHKCSKFKPDRCH 130
+++LW F K++ DPG + T E QR ++ER G A +C C +P R
Sbjct 386 SLLLWVCFLKSWKGDPGIIRPTRE-QRFKTIIELSERGGIGFEPASFCSGCLVRRPIRSK 444
Query 131 HSSNSGRCVLKMDHYCPWVANDVGFFNYKFF--FLTLLYSGATLIFVCFTMAATVRTSID 188
H S RCV + DH+CPWV N +G N+ +F FL +L LI + + + ++
Sbjct 445 HCSVCDRCVARFDHHCPWVGNCIGLKNHSYFMGFLWML-----LIMCAWMLYGGSKYYVN 499
Query 189 DSNISFEQVFFTF-------------LGTALSFFLLAIVVPFCIFHLVLITSNCTTIEFC 235
N+ F+ +G AL I++ C + V I TT E
Sbjct 500 QCNVRFDDFLGAMRAIGNCDAWVGWVMGNALLHMSWVILLTICQTYQV-ICLGMTTNERM 558
Query 236 EKRR------TGRPNPYDVGVAENL 254
+ R G +P+ G +NL
Sbjct 559 NRGRYRHFQAKGGHSPFTRGPIQNL 583
> YDR459c
Length=374
Score = 64.7 bits (156), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 47/189 (24%), Positives = 80/189 (42%), Gaps = 37/189 (19%)
Query 117 YCHKCSKFKPDRCHHSSNSGRCVLKMDHYCPWVANDVGFFNYKFFFLTLLYSGATLIFVC 176
+C +C K +R HHSS G C+ + DHYC W+ +G NY+ F Y L+ +
Sbjct 130 WCSECQSLKMERTHHSSELGHCIPRFDHYCMWIGTVIGRDNYRLFVQFAAYFSTLLLIMW 189
Query 177 FTMAATVRTSIDDSNISFE-----QVFFTFLGTALSFFLLAIVVPFCIFHLVLITSNCTT 231
++ +R I N ++ + T + L + L A ++ IF++ + N T+
Sbjct 190 VSICVYIRI-ITQHNHNYSPNLNANIISTLVFAILGWLLTASLLASSIFYM---SQNKTS 245
Query 232 IE---------------FC------------EKRRTGRPNPYD-VGVAENLKQALGDQPL 263
+E FC E R+ + +D + N+K +G L
Sbjct 246 LEAIIDSKRKKFGTRKIFCYYSEANKLRFVVEFDRSEFHSFWDKKSILANIKDFMGSNIL 305
Query 264 LWPIPVGGP 272
+W IP+G P
Sbjct 306 MWIIPLGKP 314
> Hs22060186
Length=632
Score = 63.9 bits (154), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 53/216 (24%), Positives = 93/216 (43%), Gaps = 28/216 (12%)
Query 64 LHFFVAH----AGMAMVLWSFYKTYSTDPGRVPDTDEWQRQPPPSLITERKRDGGARYCH 119
L+F H A + ++F K++ +DPG + T+E Q++ + E + +C
Sbjct 382 LNFLFIHLPFLANSVALFYNFGKSWKSDPGIIKATEE-QKKKTIVELAETGSLDLSIFCS 440
Query 120 KCSKFKPDRCHHSSNSGRCVLKMDHYCPWVANDVGFFNYKFF---FLTLLYSGATLIFVC 176
C KP R H RC+ K DH+CPWV N VG N+++F LL+ +I+ C
Sbjct 441 TCLIRKPVRSKHCGVCNRCIAKFDHHCPWVGNCVGAGNHRYFMGYLFFLLFMICWMIYGC 500
Query 177 FT---MAATVRTSIDDSNISFEQV-------FFTFLGTALSFFLLAIVVPFCIFHLVLIT 226
+ + + D Q+ F+ FL + F +A+++ ++ + +
Sbjct 501 ISYWGLHCETTYTKDGFWTYITQIATCSPWMFWMFLNSVFHFMWVAVLLMCQMYQISCL- 559
Query 227 SNCTTIEFCEKRR--------TGRPNPYDVGVAENL 254
TT E RR T +P++ G N+
Sbjct 560 -GITTNERMNARRYKHFKVTTTSIESPFNHGCVRNI 594
> CE06958
Length=306
Score = 62.8 bits (151), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 53/189 (28%), Positives = 80/189 (42%), Gaps = 34/189 (17%)
Query 99 RQPPPSLITERKRDGGARYCHKCSKFKPDRCHHSSNSGRCVLKMDHYCPWVANDVGFFNY 158
R PP + G +C KC+ +K D HH S +CVL MDH+C W+ VG N+
Sbjct 89 RTIPP---VANPGEEGDSFCSKCNYWKSDNAHHCSVCEKCVLGMDHHCIWINQCVGLHNH 145
Query 159 KFFFL---TLLYSGATLIFVCF----------TMAATVRTSI-----------DDSNISF 194
+ FFL L + AT+I + + T T+I D +
Sbjct 146 RHFFLFIANLTLAAATIIIAGYQSFSDHLFLESSQTTYCTTILEHAPLQDIICDYDGFAR 205
Query 195 EQVFFTFLGTALSFFLLAIVVPFCIFHLVLITSNCTTIEFCE----KRRTGRPNPYDVGV 250
V F +L LS LL +V +++ LI+ CT I++ + K+ T + G
Sbjct 206 TSVVFCYL---LSGILLVMVGGLTSWNIYLISIGCTYIDYLKLTGSKKNTSARKRLNKGF 262
Query 251 AENLKQALG 259
N + LG
Sbjct 263 KANWRNFLG 271
> CE12430
Length=447
Score = 62.8 bits (151), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 36/111 (32%), Positives = 57/111 (51%), Gaps = 19/111 (17%)
Query 88 PGRVPDTDEWQRQPPPSLITERKRDGGARYCHKCSKFKPDRCHHSSNSGRCVLKMDHYCP 147
PG VP W+ P + E+K ++C C+ FK R HH S RC +KMDH+CP
Sbjct 87 PGYVPRG--WR---PENAADEKK----LQFCVPCNGFKVPRSHHCSKCDRCCMKMDHHCP 137
Query 148 WVANDVGFFNYKFFFLTLLYSGATLIFVCFTMAATVRTSIDDSNISFEQVF 198
W+ N VG N+++F L F+ F++ + ++I D + + +F
Sbjct 138 WINNCVGHRNHQYF----------LRFLFFSVVGCIHSTIIDGSALYHAIF 178
> 7291055
Length=366
Score = 62.4 bits (150), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 52/190 (27%), Positives = 82/190 (43%), Gaps = 20/190 (10%)
Query 76 VLWSFYKTYSTDPGRVP------DTDEWQRQPPPSLITERKRDG---GARYCHKCSKFKP 126
VL +F DPG +P D +E R P L + +G ++C C ++P
Sbjct 66 VLANFTLATFMDPGIIPKASPDEDCEEELRAP---LYKNAEINGITVKMKWCVTCKFYRP 122
Query 127 DRCHHSSNSGRCVLKMDHYCPWVANDVGFFNYK---FFFLTLLYSGATLIFVCFTMAATV 183
RC H S C+ DH+CPWV N +G NY+ FF ++L ++ +C +
Sbjct 123 PRCSHCSVCNHCIETFDHHCPWVNNCIGRRNYRFFFFFLVSLSIHMLSIFSLCLVYVLKI 182
Query 184 RTSIDDSNISFEQVFFTFLGTALSFFLLAIVVPFCIFHLVLITSNCTTIEFCEKRRTGRP 243
+I D+ + L T L+ + + FH+VL++ TT E + G
Sbjct 183 MPNIKDT-APIVAIILMGLVTILAIPIFGLTG----FHMVLVSRGRTTNEQVTGKFKGGY 237
Query 244 NPYDVGVAEN 253
NP+ G N
Sbjct 238 NPFSRGCWHN 247
> 7294979
Length=435
Score = 62.0 bits (149), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 59/230 (25%), Positives = 92/230 (40%), Gaps = 47/230 (20%)
Query 77 LWSFYKTYSTDPGRVPDTDEWQRQPPPSLITERKRDGGARYCHKCSKFKPDRCHHSSNSG 136
L++F ++ PG VP +W P L ++ ++C +C+ +K R HH
Sbjct 62 LYNFIRSLMVGPGFVPL--KWH----PQLTKDKM---FLQFCTRCNGYKAPRSHHCRRCN 112
Query 137 RCVLKMDHYCPWVANDVGFFN------YKFFFLTLLYSGATLIFVCFTMAATVRTSIDDS 190
RCV+KMDH+CPW+ VG+ N + FF++ G +I R I
Sbjct 113 RCVMKMDHHCPWINTCVGWSNQDSFVYFLLFFMSGSIHGGIIIVSAVIRGIKKRWLIRYG 172
Query 191 NISFEQVFFTFLGTALSFFLLAIVVPFCIFHLVL-------ITSNCTTIE--FCEK---R 238
V T F L +++ + + L I N T IE +K R
Sbjct 173 LRHMATVHLTQTNLLACVFSLGVIMGTVLASIKLLYMQMKSILKNQTEIENWIVKKAAFR 232
Query 239 RTGRPN--------PYDVGVAENLKQALGDQPLLWPIPVGGPSGDGISFP 280
R P PY++G N+++ V +GDGIS+P
Sbjct 233 RNAYPRKGIKPFVYPYNLGWKTNMRE------------VFFSTGDGISWP 270
> SPAC2F7.10
Length=642
Score = 59.3 bits (142), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 33/102 (32%), Positives = 54/102 (52%), Gaps = 2/102 (1%)
Query 80 FYKTYSTDPGRVPDTDEW-QRQPPPSLITERKRDGGARYCHKCSKFKPDRCHHSSNSGRC 138
+ +T +PG V QR+ S + ++ + YC KC + KP R +H RC
Sbjct 363 YVRTAFQNPGYVDKIGAVVQRREEISKLLDKDLFNQSHYCLKCFQVKPPRSYHCGACKRC 422
Query 139 VLKMDHYCPWVANDVGFFNYKFFFLTLLYSGATLIFVCFTMA 180
+ + DH+CPW N VG N++ F L +++ +TLI + F +A
Sbjct 423 INRYDHHCPWTGNCVGARNHRTFLL-FVFTLSTLIPIYFYVA 463
> At3g56920
Length=319
Score = 58.9 bits (141), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 54/231 (23%), Positives = 90/231 (38%), Gaps = 50/231 (21%)
Query 56 RGAIAIGALH-FFVAHAGMAMVLWSF------YKTYSTDPGRVPDTDEWQRQPPPSLITE 108
R A I H FF + + +L +F + T S DPG +P + P + T+
Sbjct 53 RMAYLISHRHPFFHSLTLIGAILLTFMAFTFLFLTSSRDPGIIPRNKQVSEAEIPDVTTQ 112
Query 109 ---------------RKRDG-------GARYCHKCSKFKPDRCHHSSNSGRCVLKMDHYC 146
R +D ++C C ++P R H S CV + DH+C
Sbjct 113 STEWVTSKLGSVKLPRTKDVMVNGFTVKVKFCDTCQLYRPPRAFHCSICNNCVQRFDHHC 172
Query 147 PWVANDVGFFNYKFFFLTLLYSGATLIFV-CFTMAATVRTSIDDSNISFEQVFFTFLGTA 205
PWV + NY FF L S I+V F+ + ++ + F+ L
Sbjct 173 PWVGQCIALRNYPFFVCFLSCSTLLCIYVFVFSWVSMLKVHGE---------FYVVLADD 223
Query 206 LSFFLLAIVVPFCIFHLVLITSNCTTIEFCEKRRTGRPNPYDVGVAENLKQ 256
L +L ++ +C +++ C + ++ NPY G+ EN K+
Sbjct 224 L---ILGVLGLYC-----FVSTTCENFRYHYDKKE---NPYRKGILENFKE 263
Lambda K H
0.329 0.142 0.468
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6233576096
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40