bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0628_orf2
Length=259
Score E
Sequences producing significant alignments: (Bits) Value
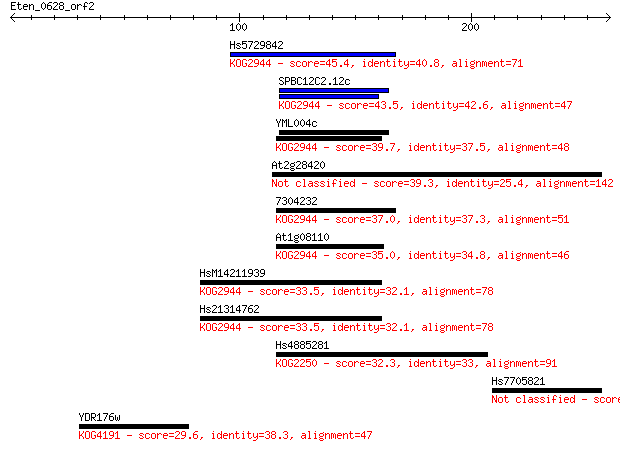
Hs5729842 45.4 1e-04
SPBC12C2.12c 43.5 5e-04
YML004c 39.7 0.006
At2g28420 39.3 0.009
7304232 37.0 0.040
At1g08110 35.0 0.14
HsM14211939 33.5 0.41
Hs21314762 33.5 0.49
Hs4885281 32.3 1.0
Hs7705821 29.6 6.3
YDR176w 29.6 7.4
> Hs5729842
Length=184
Score = 45.4 bits (106), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 29/71 (40%), Positives = 38/71 (53%), Gaps = 0/71 (0%)
Query 96 LSDNSSPSFRFDTLPLQKLPVLHHIEISVADVKKTLAFYENVLGMTLIDKKIAQGFGFTL 155
L+D ++ S D P K +L + V D KK+L FY VLGMTLI K F+L
Sbjct 11 LTDEAALSCCSDADPSTKDFLLQQTMLRVKDPKKSLDFYTRVLGMTLIQKCDFPIMKFSL 70
Query 156 YFLSLEAPTNI 166
YFL+ E +I
Sbjct 71 YFLAYEDKNDI 81
> SPBC12C2.12c
Length=302
Score = 43.5 bits (101), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 20/47 (42%), Positives = 29/47 (61%), Gaps = 0/47 (0%)
Query 117 LHHIEISVADVKKTLAFYENVLGMTLIDKKIAQGFGFTLYFLSLEAP 163
L+H I V D+ K+L FY V GM LID+ + + F+L FL+ + P
Sbjct 12 LNHTMIRVKDLDKSLKFYTEVFGMKLIDQWVFEENEFSLSFLAFDGP 58
Score = 31.6 bits (70), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 18/43 (41%), Positives = 25/43 (58%), Gaps = 1/43 (2%)
Query 117 LHHIEISVADVKKTLAFYENVLGMTLIDKKIAQGFGFTLYFLS 159
+H + V D + ++AFYE LGM +IDK FT YFL+
Sbjct 167 FNHTMVRVKDPEPSIAFYEK-LGMKVIDKADHPNGKFTNYFLA 208
> YML004c
Length=326
Score = 39.7 bits (91), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 18/47 (38%), Positives = 26/47 (55%), Gaps = 0/47 (0%)
Query 117 LHHIEISVADVKKTLAFYENVLGMTLIDKKIAQGFGFTLYFLSLEAP 163
+H I + + ++L FY+NVLGM L+ + FTLYFL P
Sbjct 183 FNHTMIRIKNPTRSLEFYQNVLGMKLLRTSEHESAKFTLYFLGYGVP 229
Score = 36.2 bits (82), Expect = 0.069, Method: Compositional matrix adjust.
Identities = 17/45 (37%), Positives = 26/45 (57%), Gaps = 0/45 (0%)
Query 116 VLHHIEISVADVKKTLAFYENVLGMTLIDKKIAQGFGFTLYFLSL 160
+L+H + V D +T+ FY GM L+ +K + F+LYFLS
Sbjct 22 LLNHTCLRVKDPARTVKFYTEHFGMKLLSRKDFEEAKFSLYFLSF 66
> At2g28420
Length=184
Score = 39.3 bits (90), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 36/144 (25%), Positives = 53/144 (36%), Gaps = 22/144 (15%)
Query 114 LPVLHHIEISVADVKKTLAFYENVLGMTLIDKKIAQGFGFTLYFLSLEAPTNISFH-YWL 172
L L+H+ DVKK+L FY VLG F+ +E P + F WL
Sbjct 18 LMALNHVSRLCKDVKKSLEFYTKVLG-----------------FVEIERPASFDFDGAWL 60
Query 173 WTQRFSTICIKVHTDGTPVKDYEDLQPHECGFLGISFLCAESEG-TTLLKRIKSYNIRVD 231
+ ++ D + L P + ISF C + E LK +K I+
Sbjct 61 FNYGVGIHLVQAKDQDKLPSDTDHLDPMDN---HISFQCEDMEALEKRLKEVKVKYIKRT 117
Query 232 TVDDEVYGQQVLLLRDPQNIPIRI 255
D++ L DP + I
Sbjct 118 VGDEKDAAIDQLFFNDPDGFMVEI 141
> 7304232
Length=176
Score = 37.0 bits (84), Expect = 0.040, Method: Compositional matrix adjust.
Identities = 19/51 (37%), Positives = 27/51 (52%), Gaps = 0/51 (0%)
Query 116 VLHHIEISVADVKKTLAFYENVLGMTLIDKKIAQGFGFTLYFLSLEAPTNI 166
+ + D +K+L FY VLGMTL+ K F+LYFL E T++
Sbjct 27 LFQQTMYRIKDPRKSLPFYTGVLGMTLLVKLDFPEAKFSLYFLGYENATDV 77
> At1g08110
Length=185
Score = 35.0 bits (79), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 16/46 (34%), Positives = 24/46 (52%), Gaps = 0/46 (0%)
Query 116 VLHHIEISVADVKKTLAFYENVLGMTLIDKKIAQGFGFTLYFLSLE 161
++ + D K +L FY VLGM+L+ + F+LYFL E
Sbjct 27 IMQQTMFRIKDPKASLDFYSRVLGMSLLKRLDFSEMKFSLYFLGYE 72
> HsM14211939
Length=176
Score = 33.5 bits (75), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 25/78 (32%), Positives = 40/78 (51%), Gaps = 5/78 (6%)
Query 83 LPAPTPEEVKAVELSDNSSPSFRFDTLPLQKLPVLHHIEISVADVKKTLAFYENVLGMTL 142
L AP P V S S P + T + L L+H+ I+V D++K AFY+N+LG +
Sbjct 19 LQAPIP----TVRASSTSQPLDQV-TGSVWNLGRLNHVAIAVPDLEKAAAFYKNILGAQV 73
Query 143 IDKKIAQGFGFTLYFLSL 160
+ G ++ F++L
Sbjct 74 SEAVPLPEHGVSVVFVNL 91
> Hs21314762
Length=176
Score = 33.5 bits (75), Expect = 0.49, Method: Compositional matrix adjust.
Identities = 25/78 (32%), Positives = 40/78 (51%), Gaps = 5/78 (6%)
Query 83 LPAPTPEEVKAVELSDNSSPSFRFDTLPLQKLPVLHHIEISVADVKKTLAFYENVLGMTL 142
L AP P V S S P + T + L L+H+ I+V D++K AFY+N+LG +
Sbjct 19 LQAPIP----TVRASSTSQPLDQV-TGSVWNLGRLNHVAIAVPDLEKAAAFYKNILGAQV 73
Query 143 IDKKIAQGFGFTLYFLSL 160
+ G ++ F++L
Sbjct 74 SEAVPLPEHGVSVVFVNL 91
> Hs4885281
Length=558
Score = 32.3 bits (72), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 30/102 (29%), Positives = 40/102 (39%), Gaps = 28/102 (27%)
Query 116 VLHHIEISVADVKKTLAFYENVLGMT--LIDKK-IAQGFGFTLYFLSLEAPTNISFHYWL 172
V H IE + + A Y ++LGMT DK + QGFG N+ H
Sbjct 276 VFHGIENFINE-----ASYMSILGMTPGFGDKTFVVQGFG------------NVGLHSMR 318
Query 173 WTQRFSTICIKV--------HTDGTPVKDYEDLQPHECGFLG 206
+ RF CI V + DG K+ ED + LG
Sbjct 319 YLHRFGAKCIAVGESDGSIWNPDGIDPKELEDFKLQHGSILG 360
> Hs7705821
Length=104
Score = 29.6 bits (65), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 15/47 (31%), Positives = 26/47 (55%), Gaps = 3/47 (6%)
Query 209 FLCAESEGTTLLKRIKSYNIRVDTVDDEVYGQQVLLLRDPQNIPIRI 255
FLC E+ +LK++ Y + ++++Y Q LLL DP +R+
Sbjct 46 FLCMETHYEKVLKKVSKY---IQEQNEKIYAPQGLLLTDPIERGLRV 89
> YDR176w
Length=702
Score = 29.6 bits (65), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 18/47 (38%), Positives = 21/47 (44%), Gaps = 0/47 (0%)
Query 31 PNQGPPDTTVGSKDEADTTHPYPFPSPQSSCCSLSVPGSVASSFFSN 77
P GP T V KDE D Y PSP S+ S +P A+ N
Sbjct 343 PKLGPLYTDVWFKDENDKNSAYKKPSPYSNDASTILPKKSANELDDN 389
Lambda K H
0.316 0.135 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5436237798
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40