bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0725_orf1
Length=332
Score E
Sequences producing significant alignments: (Bits) Value
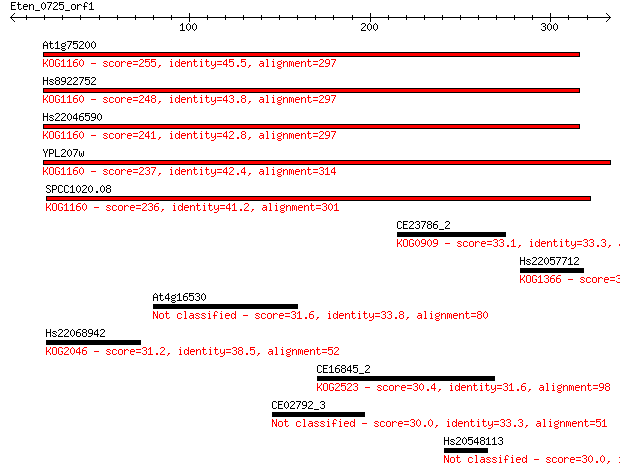
At1g75200 255 9e-68
Hs8922752 248 1e-65
Hs22046590 241 1e-63
YPL207w 237 2e-62
SPCC1020.08 236 4e-62
CE23786_2 33.1 0.96
Hs22057712 31.6 2.5
At4g16530 31.6 2.6
Hs22068942 31.2 3.1
CE16845_2 30.4 5.6
CE02792_3 30.0 6.7
Hs20548113 30.0 8.1
> At1g75200
Length=654
Score = 255 bits (651), Expect = 9e-68, Method: Compositional matrix adjust.
Identities = 135/298 (45%), Positives = 178/298 (59%), Gaps = 41/298 (13%)
Query 19 RELTGLPGLRDDFYYEVKDQILHCALSLVGEPIMYPRINELLHQMHQRRISTFLVTNAQH 78
+++ G+PG+ + E + HCALSLVGEPIMYP IN L+ ++H RRISTFLVTNAQ
Sbjct 381 KQMKGVPGVTPEKLQEGLNP-RHCALSLVGEPIMYPEINALVDELHGRRISTFLVTNAQF 439
Query 79 PDALRALRPVTQLYLSIDAATESDLKRLDKPVHRDSWLRFLSCIDILKTKKQRTVFRLTL 138
P+ + ++P+TQLY+S+DAAT+ LK +D+P+ D W RF+ + L+ K+QRTV+RLTL
Sbjct 440 PEKILMMKPITQLYVSVDAATKESLKAIDRPLFADFWERFIDSLKALQEKQQRTVYRLTL 499
Query 139 VHGYNCILGPVGDAGASEPANNRNAASNSGSSTSNTKYAKGVTSTLDKYASLVLRGEPDM 198
V G+N T LD Y +L G+PD
Sbjct 500 VKGWN-------------------------------------TEELDAYFNLFSIGKPDF 522
Query 199 IEIKGMTFCGGLDRESLSMQNVPRHQQVLAFADGLRRALPEGLYGLASEHEHSCCVLLAN 258
IEIKG+T+CG L+M+NVP H V AF++ L G Y +A EH HSCCVLL
Sbjct 523 IEIKGVTYCGSSATSKLTMENVPWHTDVKAFSEALSLK-SNGEYEVACEHAHSCCVLLGR 581
Query 259 NKYF-YKGQWHTWIDYPKFFDLALSGDLNFSGIDYSAATPSWAIFGAEGRGFDPQQKR 315
+ F G+W TWIDY KF DL SG+ F+ DY A TPSWA++GA+ GFDP Q R
Sbjct 582 TEKFKVDGKWFTWIDYEKFHDLVASGE-PFTSTDYMAQTPSWAVYGAQEGGFDPGQLR 638
> Hs8922752
Length=732
Score = 248 bits (633), Expect = 1e-65, Method: Compositional matrix adjust.
Identities = 130/302 (43%), Positives = 175/302 (57%), Gaps = 45/302 (14%)
Query 19 RELTGLPGLRDDFYYEVKDQILHCALSLVGEPIMYPRINELLHQMHQRRISTFLVTNAQH 78
++ G+PG++ + +E + HCALSLVGEPIMYP IN L +HQ +IS+FLVTNAQ
Sbjct 458 KQFKGVPGVKAE-RFEEGMTVKHCALSLVGEPIMYPEINRFLKLLHQCKISSFLVTNAQF 516
Query 79 PDALRALRPVTQLYLSIDAATESDLKRLDKPVHRDSWLRFLSCIDILKTKKQRTVFRLTL 138
P +R L PVTQLY+S+DA+T+ LK++D+P+ +D W RFL + L K+QRTV+RLTL
Sbjct 517 PAEIRNLEPVTQLYVSVDASTKDSLKKIDRPLFKDFWQRFLDSLKALAVKQQRTVYRLTL 576
Query 139 VHGYNCILGPVGDAGASEPANNRNAASNSGSSTSNTKYAKGVTSTLDKYASLVLRGEPDM 198
V +N L YA LV G PD
Sbjct 577 VKAWN-------------------------------------VDELQAYAQLVSLGNPDF 599
Query 199 IEIKGMTFCGGLDRESLSMQNVPRHQQVLAFADGLRRALPEGLYGLASEHEHSCCVLLAN 258
IE+KG+T+CG SL+M +VP H++V+ F L +PE Y +A EHEHS C+L+A+
Sbjct 600 IEVKGVTYCGESSASSLTMAHVPWHEEVVQFVRELVDLIPE--YEIACEHEHSNCLLIAH 657
Query 259 NKYFYKGQWHTWIDYPKFFDLALS-----GDLNFSGIDYSAATPSWAIFGAEGRGFDPQQ 313
K+ G+W TWI+Y +F +L G FS DY A TP WA+FGA RGFDP+
Sbjct 658 RKFKIGGEWWTWINYNRFQELIQEYEDSGGSKTFSAKDYMARTPHWALFGASERGFDPKD 717
Query 314 KR 315
R
Sbjct 718 TR 719
> Hs22046590
Length=294
Score = 241 bits (615), Expect = 1e-63, Method: Compositional matrix adjust.
Identities = 127/302 (42%), Positives = 172/302 (56%), Gaps = 45/302 (14%)
Query 19 RELTGLPGLRDDFYYEVKDQILHCALSLVGEPIMYPRINELLHQMHQRRISTFLVTNAQH 78
++ G+PG++ + +E + HCALSLVGEPIMYP IN L +HQ +IS+FLVTNAQ
Sbjct 20 KQFKGVPGVKAE-RFEEGMTVKHCALSLVGEPIMYPEINRFLKLLHQCKISSFLVTNAQF 78
Query 79 PDALRALRPVTQLYLSIDAATESDLKRLDKPVHRDSWLRFLSCIDILKTKKQRTVFRLTL 138
P +R L PVTQLY+S+DA+T+ LK++D+P+ +D W +FL + L K+QRTV+RL L
Sbjct 79 PAEIRNLEPVTQLYVSVDASTKDSLKKIDRPLFKDFWQQFLDSLKALAVKQQRTVYRLML 138
Query 139 VHGYNCILGPVGDAGASEPANNRNAASNSGSSTSNTKYAKGVTSTLDKYASLVLRGEPDM 198
V +N L YA LV G PD
Sbjct 139 VKAWN-------------------------------------VDELQAYAQLVSLGNPDF 161
Query 199 IEIKGMTFCGGLDRESLSMQNVPRHQQVLAFADGLRRALPEGLYGLASEHEHSCCVLLAN 258
IE+KG+T+C SL+M +VP H++V+ F L +PE Y +A EHEHS C+L+A+
Sbjct 162 IEVKGVTYCRESSASSLTMAHVPWHEEVVQFVRELVDLIPE--YEIACEHEHSNCLLIAH 219
Query 259 NKYFYKGQWHTWIDYPKFFDLALS-----GDLNFSGIDYSAATPSWAIFGAEGRGFDPQQ 313
K+ G+W TWIDY +F +L G FS DY A TP WA+FGA R FDP+
Sbjct 220 RKFKIGGEWWTWIDYNRFQELIQEYEDSGGSKTFSAKDYMARTPHWALFGANERSFDPKD 279
Query 314 KR 315
R
Sbjct 280 TR 281
> YPL207w
Length=810
Score = 237 bits (605), Expect = 2e-62, Method: Compositional matrix adjust.
Identities = 133/320 (41%), Positives = 191/320 (59%), Gaps = 47/320 (14%)
Query 19 RELTGLPGLRDDFYYEVKDQILHCALSLVGEPIMYPRINELLHQMHQRRISTFLVTNAQH 78
+++ G+PG+ + + + ++ HCALSLVGEPI+YP IN+ + +HQ+ I++FLV NAQH
Sbjct 521 KQMRGVPGVIAERFAKAF-EVRHCALSLVGEPILYPHINKFIQLLHQKGITSFLVCNAQH 579
Query 79 PDALRALRPVTQLYLSIDAATESDLKRLDKPVHRDSWLRFLSCIDILKTKK--QRTVFRL 136
P+ALR + VTQLY+SIDA T+++LK++D+P+++D W R + C++ILKT + QRTVFRL
Sbjct 580 PEALRNIVKVTQLYVSIDAPTKTELKKVDRPLYKDFWERMVECLEILKTVQNHQRTVFRL 639
Query 137 TLVHGYNCILGPVGDAGASEPANNRNAASNSGSSTSNTKYAKGVTSTLDKYASLVLRGEP 196
TLV G+N +GD A YA LV RG P
Sbjct 640 TLVKGFN-----MGDVSA--------------------------------YADLVQRGLP 662
Query 197 DMIEIKGMTFCGGLDRES--LSMQNVPRHQQVLAFADGLRRALP-EGL-YGLASEHEHSC 252
IE+KG TF G D L+MQN+P +++ + F L GL Y LA+EH HS
Sbjct 663 GFIEVKGATFSGSSDGNGNPLTMQNIPFYEECVKFVKAFTTELQRRGLHYDLAAEHAHSN 722
Query 253 CVLLANNKYFYKGQWHTWIDYPKFFDLALSGDLNFSGIDYSAATPSWAIFGAEGRGFDPQ 312
C+L+A+ K+ G+WHT ID+ KFF L SG +F+ +DY TP WA+FG GF P
Sbjct 723 CLLIADTKFKINGEWHTHIDFDKFFVLLNSGK-DFTYMDYLEKTPEWALFG--NGGFAPG 779
Query 313 QKRVFGKGRRGKELELSQQT 332
RV+ K ++ + E + T
Sbjct 780 NTRVYRKDKKKQNKENQETT 799
> SPCC1020.08
Length=688
Score = 236 bits (603), Expect = 4e-62, Method: Compositional matrix adjust.
Identities = 124/307 (40%), Positives = 178/307 (57%), Gaps = 47/307 (15%)
Query 21 LTGLPGLRDDFYYEVKDQILHCALSLVGEPIMYPRINELLHQMHQRRISTFLVTNAQHPD 80
+ G+PG+ D Y E ++ HCALSLVGEPI YP INE + +H+R IS+FLVTNAQHP+
Sbjct 401 MKGVPGVLPDRYEEA-SRVRHCALSLVGEPIFYPYINEFVSMLHEREISSFLVTNAQHPE 459
Query 81 ALRALRPVTQLYLSIDAATESDLKRLDKPVHRDSWLRFLSCIDILKTKKQRTVFRLTLVH 140
ALR + VTQLY+S+DA+T+ LK +D+P+ +D W R L+C++IL+ K+QRTV+R+TLV
Sbjct 460 ALRNMGMVTQLYVSVDASTKQSLKSVDRPLFKDFWERMLTCLEILREKRQRTVYRMTLVK 519
Query 141 GYNCILGPVGDAGASEPANNRNAASNSGSSTSNTKYAKGVTSTLDKYASLVLRGEPDMIE 200
G+N + +Y L+ G P IE
Sbjct 520 GFN-------------------------------------MEQIKEYTELIRLGVPCFIE 542
Query 201 IKGMTFCGGLDRESLSMQNVPRHQQVLAFADGLRRALPEGL------YGLASEHEHSCCV 254
+KG+T+ G D+ L+M+NVP +++V+ F L + L Y +A+EH HSC +
Sbjct 543 VKGVTYSGNSDQSPLTMKNVPYYEEVIDFVKKLIEYIDIHLQDLGVRYEIAAEHAHSCSI 602
Query 255 LLANNKYFYKGQWHTWIDYPKFFDLALSGDLNFSGIDYSAATPSWAIFGAEGRGFDPQQK 314
L+A + G WHT IDYPKFF+L + +F DY A+TP +A+FG GF P+
Sbjct 603 LVAQTAFKKDGHWHTHIDYPKFFEL-IRTKKDFGPFDYMASTPDFAMFG--NGGFSPEDT 659
Query 315 RVFGKGR 321
R K +
Sbjct 660 RFHRKKK 666
> CE23786_2
Length=500
Score = 33.1 bits (74), Expect = 0.96, Method: Compositional matrix adjust.
Identities = 20/61 (32%), Positives = 29/61 (47%), Gaps = 1/61 (1%)
Query 215 LSMQNVPRHQQVLAFADGLRRALPEGLYGLA-SEHEHSCCVLLANNKYFYKGQWHTWIDY 273
+ NVPR Q F R +PE L G A +E +L + +++K Q+ TW D
Sbjct 23 VKCSNVPRSYQDEVFKALARSVMPEELVGRAMTEGPRDEKAILKDLLHWFKTQFFTWFDR 82
Query 274 P 274
P
Sbjct 83 P 83
> Hs22057712
Length=1857
Score = 31.6 bits (70), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 16/37 (43%), Positives = 22/37 (59%), Gaps = 5/37 (13%)
Query 283 GDLNFSGIDYSAATPSWAIFGAEGRGF--DPQQKRVF 317
G + ++GI Y TP+WA FG +G DPQ R+F
Sbjct 516 GHILYNGIYY---TPAWADFGKDGYDLVKDPQNNRIF 549
> At4g16530
Length=773
Score = 31.6 bits (70), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 27/94 (28%), Positives = 40/94 (42%), Gaps = 15/94 (15%)
Query 80 DALRALRPVTQLYLSIDAATESDLKRLDKPV----------HRDSWLRFLSCIDILKTKK 129
D+L ++P+ L L++ ESD+K L K V H+D W CI L + K
Sbjct 100 DSLYEIKPLLILCLTMQETKESDIKILRKIVSFVTYNVVELHKDKWDELGDCILSLASSK 159
Query 130 QRT----VFRLTLVHGYNCILGPVGDAGASEPAN 159
+ VF + L Y +G D E +N
Sbjct 160 EPVKAFHVF-IDLPPVYKSFIGKFLDKILEEASN 192
> Hs22068942
Length=311
Score = 31.2 bits (69), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 28/55 (50%), Gaps = 3/55 (5%)
Query 21 LTGLPGL--RDDFYYEVKDQILHCALSLVGEPIMYPRINELLHQMHQ-RRISTFL 72
+ GL GL DF +KD I+ CAL +P P+IN + HQ +S F+
Sbjct 38 IKGLTGLFIGPDFQKGLKDGIILCALMNKLQPGSVPKINHSMQNWHQLENLSNFI 92
> CE16845_2
Length=180
Score = 30.4 bits (67), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 31/106 (29%), Positives = 43/106 (40%), Gaps = 9/106 (8%)
Query 171 TSNTKYAKGVTSTLDKYASLVLRGEPDMIEIKGM-----TFCGGLDRESLSMQN-VPRHQ 224
T NT Y + TL KY ++ + D IK + C GL + VP+
Sbjct 69 TRNTDYIPTL-RTLHKYPFILPHQQVDKGAIKFILNGSSIMCPGLTSPGAKLTPLVPKDS 127
Query 225 QVLAFADGLRRALPEGLYGLASEHEHSCCV--LLANNKYFYKGQWH 268
V A+G + AL GL ++SE S + N Y G WH
Sbjct 128 VVAVMAEGKQHALAIGLMSMSSEEIQSVNKGNGIENLHYLNDGLWH 173
> CE02792_3
Length=235
Score = 30.0 bits (66), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 27/51 (52%), Gaps = 0/51 (0%)
Query 146 LGPVGDAGASEPANNRNAASNSGSSTSNTKYAKGVTSTLDKYASLVLRGEP 196
LG VG++G E + N + S S+++ +TS L K S +RG+P
Sbjct 75 LGFVGESGGDEMSRNDDPVVTSNGSSTDVNRGSRITSELKKKESNAVRGQP 125
> Hs20548113
Length=394
Score = 30.0 bits (66), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 13/24 (54%), Positives = 16/24 (66%), Gaps = 0/24 (0%)
Query 241 LYGLASEHEHSCCVLLANNKYFYK 264
L G ASEHE SCC L A + + +K
Sbjct 206 LRGGASEHEESCCRLAATSLFLHK 229
Lambda K H
0.321 0.136 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7847861938
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40