bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0752_orf2
Length=205
Score E
Sequences producing significant alignments: (Bits) Value
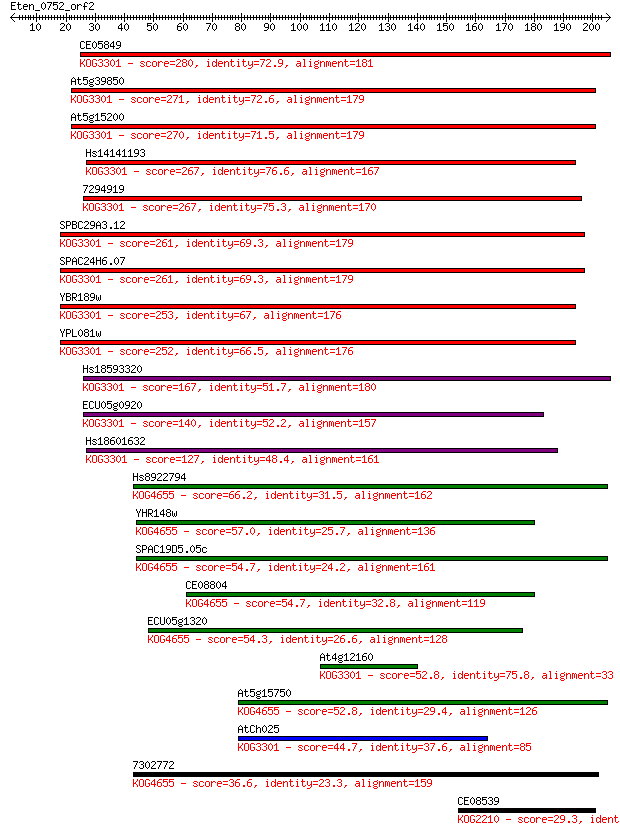
CE05849 280 1e-75
At5g39850 271 1e-72
At5g15200 270 1e-72
Hs14141193 267 9e-72
7294919 267 9e-72
SPBC29A3.12 261 6e-70
SPAC24H6.07 261 9e-70
YBR189w 253 3e-67
YPL081w 252 4e-67
Hs18593320 167 1e-41
ECU05g0920 140 2e-33
Hs18601632 127 1e-29
Hs8922794 66.2 4e-11
YHR148w 57.0 3e-08
SPAC19D5.05c 54.7 1e-07
CE08804 54.7 1e-07
ECU05g1320 54.3 2e-07
At4g12160 52.8 4e-07
At5g15750 52.8 5e-07
AtCh025 44.7 1e-04
7302772 36.6 0.038
CE08539 29.3 5.2
> CE05849
Length=189
Score = 280 bits (717), Expect = 1e-75, Method: Compositional matrix adjust.
Identities = 132/181 (72%), Positives = 162/181 (89%), Gaps = 0/181 (0%)
Query 25 QSKTARNPKRPFEKERLDQELRLLGEYGLKNKREVWRVQYALAKIRSAARELLTLEEKDP 84
QSK ++P+RPFEKERLDQEL+L+G +GLKNKREVWRV+Y LAK+R AARELLTLE+KDP
Sbjct 8 QSKVTKSPRRPFEKERLDQELKLIGTFGLKNKREVWRVKYTLAKVRKAARELLTLEDKDP 67
Query 85 RRIFQGTALLRRMVRLGLLSEGEQKLDYVLGLTVQKFLERRLQTRVFKLGLAKSIHHARC 144
+R+F+G ALLRR+V++G+L E + KLDYVLGL V+ FLERRLQT+VFKLGLAKSIHHAR
Sbjct 68 KRLFEGNALLRRLVKIGVLDETKMKLDYVLGLKVEDFLERRLQTQVFKLGLAKSIHHARI 127
Query 145 LIRQRHIRVGKQIVDIPSFMVRVDSEKYIEFSLTSPYGGGRPGRVKRKSLRGGDDKDGDD 204
LI+Q HIRV +Q+VD+PSF+VR+DS+K+I+FSL SPYGGGRPGRVKR++LR GD GDD
Sbjct 128 LIKQHHIRVRRQVVDVPSFIVRLDSQKHIDFSLQSPYGGGRPGRVKRRTLRKGDGAGGDD 187
Query 205 E 205
E
Sbjct 188 E 188
> At5g39850
Length=197
Score = 271 bits (692), Expect = 1e-72, Method: Compositional matrix adjust.
Identities = 130/179 (72%), Positives = 157/179 (87%), Gaps = 0/179 (0%)
Query 22 YRNQSKTARNPKRPFEKERLDQELRLLGEYGLKNKREVWRVQYALAKIRSAARELLTLEE 81
YRN KT + P+RP+EKERLD EL+L+GEYGL+ KRE+WRVQY L++IR+AARELLTL+E
Sbjct 7 YRNYGKTFKKPRRPYEKERLDAELKLVGEYGLRCKRELWRVQYTLSRIRNAARELLTLDE 66
Query 82 KDPRRIFQGTALLRRMVRLGLLSEGEQKLDYVLGLTVQKFLERRLQTRVFKLGLAKSIHH 141
K+PRRIF+G ALLRRM R GLL E + KLDYVL LTV+ FLERRLQT VFK G+AKSIHH
Sbjct 67 KNPRRIFEGEALLRRMNRYGLLDETQNKLDYVLALTVENFLERRLQTIVFKSGMAKSIHH 126
Query 142 ARCLIRQRHIRVGKQIVDIPSFMVRVDSEKYIEFSLTSPYGGGRPGRVKRKSLRGGDDK 200
AR LIRQRHIRVG+Q+V+IPSFMVRV+S+K+++FSLTSP+GGGRPGRVKR++ R G K
Sbjct 127 ARVLIRQRHIRVGRQLVNIPSFMVRVESQKHVDFSLTSPFGGGRPGRVKRRNERAGAKK 185
> At5g15200
Length=198
Score = 270 bits (690), Expect = 1e-72, Method: Compositional matrix adjust.
Identities = 128/179 (71%), Positives = 157/179 (87%), Gaps = 0/179 (0%)
Query 22 YRNQSKTARNPKRPFEKERLDQELRLLGEYGLKNKREVWRVQYALAKIRSAARELLTLEE 81
YRN KT + P+RP+EKERLD EL+L+GEYGL+NKRE+WRVQY+L++IR+AAR+LLTL+E
Sbjct 7 YRNYGKTFKGPRRPYEKERLDSELKLVGEYGLRNKRELWRVQYSLSRIRNAARDLLTLDE 66
Query 82 KDPRRIFQGTALLRRMVRLGLLSEGEQKLDYVLGLTVQKFLERRLQTRVFKLGLAKSIHH 141
K PRRIF+G ALLRRM R GLL E + KLDYVL LTV+ FLERRLQT VFK G+AKSIHH
Sbjct 67 KSPRRIFEGEALLRRMNRYGLLDESQNKLDYVLALTVENFLERRLQTIVFKSGMAKSIHH 126
Query 142 ARCLIRQRHIRVGKQIVDIPSFMVRVDSEKYIEFSLTSPYGGGRPGRVKRKSLRGGDDK 200
+R LIRQRHIRVGKQ+V+IPSFMVR+DS+K+I+F+LTSP+GGGRPGRVKR++ + K
Sbjct 127 SRVLIRQRHIRVGKQLVNIPSFMVRLDSQKHIDFALTSPFGGGRPGRVKRRNEKSASKK 185
> Hs14141193
Length=194
Score = 267 bits (683), Expect = 9e-72, Method: Compositional matrix adjust.
Identities = 128/167 (76%), Positives = 153/167 (91%), Gaps = 0/167 (0%)
Query 27 KTARNPKRPFEKERLDQELRLLGEYGLKNKREVWRVQYALAKIRSAARELLTLEEKDPRR 86
KT P+RPFEK RLDQEL+L+GEYGL+NKREVWRV++ LAKIR AARELLTL+EKDPRR
Sbjct 11 KTYVTPRRPFEKSRLDQELKLIGEYGLRNKREVWRVKFTLAKIRKAARELLTLDEKDPRR 70
Query 87 IFQGTALLRRMVRLGLLSEGEQKLDYVLGLTVQKFLERRLQTRVFKLGLAKSIHHARCLI 146
+F+G ALLRR+VR+G+L EG+ KLDY+LGL ++ FLERRLQT+VFKLGLAKSIHHAR LI
Sbjct 71 LFEGNALLRRLVRIGVLDEGKMKLDYILGLKIEDFLERRLQTQVFKLGLAKSIHHARVLI 130
Query 147 RQRHIRVGKQIVDIPSFMVRVDSEKYIEFSLTSPYGGGRPGRVKRKS 193
RQRHIRV KQ+V+IPSF+VR+DS+K+I+FSL SPYGGGRPGRVKRK+
Sbjct 131 RQRHIRVRKQVVNIPSFIVRLDSQKHIDFSLRSPYGGGRPGRVKRKN 177
> 7294919
Length=195
Score = 267 bits (683), Expect = 9e-72, Method: Compositional matrix adjust.
Identities = 128/170 (75%), Positives = 154/170 (90%), Gaps = 0/170 (0%)
Query 26 SKTARNPKRPFEKERLDQELRLLGEYGLKNKREVWRVQYALAKIRSAARELLTLEEKDPR 85
SKT P+RP+EK RLDQEL+++GEYGL+NKREVWRV+YALAKIR AARELLTL+EKD +
Sbjct 11 SKTYVTPRRPYEKARLDQELKIIGEYGLRNKREVWRVKYALAKIRKAARELLTLDEKDEK 70
Query 86 RIFQGTALLRRMVRLGLLSEGEQKLDYVLGLTVQKFLERRLQTRVFKLGLAKSIHHARCL 145
R+FQG ALLRR+VR+G+L E KLDYVLGL ++ FLERRLQT+VFKLGLAKSIHHAR L
Sbjct 71 RLFQGNALLRRLVRIGVLDESRMKLDYVLGLKIEDFLERRLQTQVFKLGLAKSIHHARVL 130
Query 146 IRQRHIRVGKQIVDIPSFMVRVDSEKYIEFSLTSPYGGGRPGRVKRKSLR 195
IRQRHIRV KQ+V+IPSF+VR+DS+K+I+FSL SP+GGGRPGRVKRK+L+
Sbjct 131 IRQRHIRVRKQVVNIPSFVVRLDSQKHIDFSLKSPFGGGRPGRVKRKNLK 180
> SPBC29A3.12
Length=192
Score = 261 bits (668), Expect = 6e-70, Method: Compositional matrix adjust.
Identities = 124/179 (69%), Positives = 152/179 (84%), Gaps = 0/179 (0%)
Query 18 MPKSYRNQSKTARNPKRPFEKERLDQELRLLGEYGLKNKREVWRVQYALAKIRSAARELL 77
MP + R QSKT + P+RPFE RLD EL+L GEYGL+NK E+WRV L+KIR AARELL
Sbjct 1 MPSAPRKQSKTYKVPRRPFESARLDAELKLAGEYGLRNKHEIWRVALTLSKIRRAARELL 60
Query 78 TLEEKDPRRIFQGTALLRRMVRLGLLSEGEQKLDYVLGLTVQKFLERRLQTRVFKLGLAK 137
TL+EKDP+R+F+G A++RR+VRLG+L E KLDYVL L ++ FLERRLQT+VFKLGLAK
Sbjct 61 TLDEKDPKRLFEGNAIIRRLVRLGILDESRMKLDYVLALRIEDFLERRLQTQVFKLGLAK 120
Query 138 SIHHARCLIRQRHIRVGKQIVDIPSFMVRVDSEKYIEFSLTSPYGGGRPGRVKRKSLRG 196
SIHHAR LI QRHIRVGKQIV++PSF+VR+D++K+I+F+L+SPYGGGRPGR KRK LR
Sbjct 121 SIHHARVLIFQRHIRVGKQIVNVPSFVVRLDAQKHIDFALSSPYGGGRPGRCKRKRLRS 179
> SPAC24H6.07
Length=191
Score = 261 bits (666), Expect = 9e-70, Method: Compositional matrix adjust.
Identities = 124/179 (69%), Positives = 152/179 (84%), Gaps = 0/179 (0%)
Query 18 MPKSYRNQSKTARNPKRPFEKERLDQELRLLGEYGLKNKREVWRVQYALAKIRSAARELL 77
MP + R QSKT + P+RPFE RLD EL+L GEYGL+NK E+WRV L+KIR AARELL
Sbjct 1 MPSAPRKQSKTYKVPRRPFESARLDAELKLAGEYGLRNKHEIWRVALTLSKIRRAARELL 60
Query 78 TLEEKDPRRIFQGTALLRRMVRLGLLSEGEQKLDYVLGLTVQKFLERRLQTRVFKLGLAK 137
TL+EKDP+R+F+G A++RR+VRLG+L E KLDYVL L ++ FLERRLQT+VFKLGLAK
Sbjct 61 TLDEKDPKRLFEGNAIIRRLVRLGILDETRMKLDYVLALRIEDFLERRLQTQVFKLGLAK 120
Query 138 SIHHARCLIRQRHIRVGKQIVDIPSFMVRVDSEKYIEFSLTSPYGGGRPGRVKRKSLRG 196
SIHHAR LI QRHIRVGKQIV++PSF+VR+D++K+I+F+L+SPYGGGRPGR KRK LR
Sbjct 121 SIHHARVLIFQRHIRVGKQIVNVPSFVVRLDTQKHIDFALSSPYGGGRPGRCKRKRLRS 179
> YBR189w
Length=195
Score = 253 bits (645), Expect = 3e-67, Method: Compositional matrix adjust.
Identities = 118/176 (67%), Positives = 151/176 (85%), Gaps = 0/176 (0%)
Query 18 MPKSYRNQSKTARNPKRPFEKERLDQELRLLGEYGLKNKREVWRVQYALAKIRSAARELL 77
MP++ R SKT PKRP+E RLD EL+L GE+GLKNKRE++R+ + L+KIR AAR+LL
Sbjct 1 MPRAPRTYSKTYSTPKRPYESSRLDAELKLAGEFGLKNKREIYRISFQLSKIRRAARDLL 60
Query 78 TLEEKDPRRIFQGTALLRRMVRLGLLSEGEQKLDYVLGLTVQKFLERRLQTRVFKLGLAK 137
T +EKDP+R+F+G AL+RR+VR+G+LSE ++KLDYVL L V+ FLERRLQT+V+KLGLAK
Sbjct 61 TRDEKDPKRLFEGNALIRRLVRVGVLSEDKKKLDYVLALKVEDFLERRLQTQVYKLGLAK 120
Query 138 SIHHARCLIRQRHIRVGKQIVDIPSFMVRVDSEKYIEFSLTSPYGGGRPGRVKRKS 193
S+HHAR LI QRHI VGKQIV+IPSFMVR+DSEK+I+F+ TSP+GG RPGRV R++
Sbjct 121 SVHHARVLITQRHIAVGKQIVNIPSFMVRLDSEKHIDFAPTSPFGGARPGRVARRN 176
> YPL081w
Length=197
Score = 252 bits (643), Expect = 4e-67, Method: Compositional matrix adjust.
Identities = 117/176 (66%), Positives = 151/176 (85%), Gaps = 0/176 (0%)
Query 18 MPKSYRNQSKTARNPKRPFEKERLDQELRLLGEYGLKNKREVWRVQYALAKIRSAARELL 77
MP++ R SKT PKRP+E RLD EL+L GE+GLKNK+E++R+ + L+KIR AAR+LL
Sbjct 1 MPRAPRTYSKTYSTPKRPYESSRLDAELKLAGEFGLKNKKEIYRISFQLSKIRRAARDLL 60
Query 78 TLEEKDPRRIFQGTALLRRMVRLGLLSEGEQKLDYVLGLTVQKFLERRLQTRVFKLGLAK 137
T +EKDP+R+F+G AL+RR+VR+G+LSE ++KLDYVL L V+ FLERRLQT+V+KLGLAK
Sbjct 61 TRDEKDPKRLFEGNALIRRLVRVGVLSEDKKKLDYVLALKVEDFLERRLQTQVYKLGLAK 120
Query 138 SIHHARCLIRQRHIRVGKQIVDIPSFMVRVDSEKYIEFSLTSPYGGGRPGRVKRKS 193
S+HHAR LI QRHI VGKQIV+IPSFMVR+DSEK+I+F+ TSP+GG RPGRV R++
Sbjct 121 SVHHARVLITQRHIAVGKQIVNIPSFMVRLDSEKHIDFAPTSPFGGARPGRVARRN 176
> Hs18593320
Length=168
Score = 167 bits (423), Expect = 1e-41, Method: Compositional matrix adjust.
Identities = 93/182 (51%), Positives = 116/182 (63%), Gaps = 28/182 (15%)
Query 26 SKTARNPKRPFEKERLDQELRLLGEYGLKNKREVWRVQYALAKIRSAARELLTLEEKDPR 85
+T +RPFEK LDQ+L+L+GEYGL+NK EVWRV++ L KI AARELLTL+EKDPR
Sbjct 10 CQTYVTLQRPFEKPHLDQKLKLIGEYGLQNKCEVWRVKFTLVKIHKAARELLTLDEKDPR 69
Query 86 RIFQGTALLRRMVRLGLLSEGEQKLDYVLGLTVQKFLERRLQTRVFKLGLAKSIHHARCL 145
R+F+G ALL+ + R LQT+VFKLGLAKSI HA L
Sbjct 70 RLFEGIALLQWL--------------------------RFLQTQVFKLGLAKSIRHAHVL 103
Query 146 IRQRHIRVGKQIVDIPSFMVRVDSEKYIEFSLTSPYGGGRPGRVKRKSLRGGDDKDG--D 203
I+Q HIRV +Q+V+I F VR+DS+K+I+FSL P G P VKRK+ G G D
Sbjct 104 IQQCHIRVREQVVNILFFTVRLDSQKHIDFSLCFPIGVANPSHVKRKNASKGQGGAGARD 163
Query 204 DE 205
DE
Sbjct 164 DE 165
> ECU05g0920
Length=184
Score = 140 bits (352), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 82/166 (49%), Positives = 104/166 (62%), Gaps = 11/166 (6%)
Query 26 SKTARNPKRPFEKERLDQELRLLGEYGLKNKREVWRVQYALAKIRSAARELLTLEEKDPR 85
SK A P+ PFEKERL +E +L+G YGLKNK E+W ++ AK + AR LLT +P
Sbjct 7 SKRASTPRDPFEKERLIREAQLVGVYGLKNKHELWVMEKIFAKDKERARILLT--STNPE 64
Query 86 RI-FQGTALLRRMVRLGLLS--------EGEQKLDYVLGLTVQKFLERRLQTRVFKLGLA 136
I G +LLRR+++ G+L E L+ VL LT+ +LERRLQ RVF GLA
Sbjct 65 DIPINGRSLLRRLMKYGILGGIDLCDKQEVVNGLNKVLDLTINHYLERRLQFRVFAAGLA 124
Query 137 KSIHHARCLIRQRHIRVGKQIVDIPSFMVRVDSEKYIEFSLTSPYG 182
KS+HHAR LI R I + QIVD+P FMVR D E IE++ S YG
Sbjct 125 KSVHHARLLIGGRCISIKDQIVDVPGFMVRADKEPLIEYNPYSRYG 170
> Hs18601632
Length=131
Score = 127 bits (319), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 78/161 (48%), Positives = 97/161 (60%), Gaps = 41/161 (25%)
Query 27 KTARNPKRPFEKERLDQELRLLGEYGLKNKREVWRVQYALAKIRSAARELLTLEEKDPRR 86
KT P+RPFEK RLDQEL+L+G
Sbjct 11 KTYVTPRRPFEKSRLDQELKLIG------------------------------------- 33
Query 87 IFQGTALLRRMVRLGLLSEGEQKLDYVLGLTVQKFLERRLQTRVFKLGLAKSIHHARCLI 146
LLRR++R+G+L EG+ KLDY+LGL ++ FLERRLQT+VFKLGLAKSIHHA LI
Sbjct 34 ----NVLLRRLLRIGVLDEGKTKLDYILGLKIEDFLERRLQTQVFKLGLAKSIHHAGVLI 89
Query 147 RQRHIRVGKQIVDIPSFMVRVDSEKYIEFSLTSPYGGGRPG 187
RQR +RV Q+ +IPSF+VR+DS+K I+ SL SPYG P
Sbjct 90 RQRRVRVRNQVANIPSFIVRLDSQKRIDLSLRSPYGVAAPA 130
> Hs8922794
Length=184
Score = 66.2 bits (160), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 51/164 (31%), Positives = 82/164 (50%), Gaps = 13/164 (7%)
Query 43 QELRLLGEYGLKNKREVWRVQYALAKIRSAARELLTLEEKDPRRIFQGTALLRRMVRLGL 102
ELR+L Y L+ + + R +R AR L L E+D R+ ALL ++ LGL
Sbjct 29 HELRVLRRYRLQRREDYTRYNQLSRAVRELARRLRDLPERDQFRVRASAALLDKLYALGL 88
Query 103 L-SEGEQKL-DYVLGLTVQKFLERRLQTRVFKLGLAKSIHHARCLIRQRHIRVGKQIVDI 160
+ + G +L D+V T F RRL T + KL +A+ + A + Q H+RVG +V
Sbjct 89 VPTRGSLELCDFV---TASSFCRRRLPTVLLKLRMAQHLQAAVAFVEQGHVRVGPDVVTD 145
Query 161 PSFMVRVDSEKYIEFSLTSPYGGGRPGRVKRKSLRGGDDKDGDD 204
P+F+V E ++ + +S ++KR L +++D D
Sbjct 146 PAFLVTRSMEDFVTWVDSS--------KIKRHVLEYNEERDDFD 181
> YHR148w
Length=183
Score = 57.0 bits (136), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 35/136 (25%), Positives = 67/136 (49%), Gaps = 0/136 (0%)
Query 44 ELRLLGEYGLKNKREVWRVQYALAKIRSAARELLTLEEKDPRRIFQGTALLRRMVRLGLL 103
+ +++ Y ++N+ + + IR A +L L DP R LL ++ +G+L
Sbjct 29 DTQVMRTYHIQNREDYHKYNRICGDIRRLANKLSLLPPTDPFRRKHEQLLLDKLYAMGVL 88
Query 104 SEGEQKLDYVLGLTVQKFLERRLQTRVFKLGLAKSIHHARCLIRQRHIRVGKQIVDIPSF 163
+ + D +TV RRL + +L +A++I A I Q H+RVG +++ P++
Sbjct 89 TTKSKISDLENKVTVSAICRRRLPVIMHRLKMAETIQDAVKFIEQGHVRVGPNLINDPAY 148
Query 164 MVRVDSEKYIEFSLTS 179
+V + E Y+ + S
Sbjct 149 LVTRNMEDYVTWVDNS 164
> SPAC19D5.05c
Length=183
Score = 54.7 bits (130), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 39/161 (24%), Positives = 71/161 (44%), Gaps = 8/161 (4%)
Query 44 ELRLLGEYGLKNKREVWRVQYALAKIRSAARELLTLEEKDPRRIFQGTALLRRMVRLGLL 103
++ ++ Y + + E + K R A L L+ DP R+ LL ++ +G+L
Sbjct 29 DVMVMRRYHISKREEYQKYNIICGKFRQLAHRLSLLDPTDPFRLQYENLLLEKLFDMGIL 88
Query 104 SEGEQKLDYVLGLTVQKFLERRLQTRVFKLGLAKSIHHARCLIRQRHIRVGKQIVDIPSF 163
+ D V RRL + KL +++ + A LI Q H+RVG ++ P++
Sbjct 89 PSKSKMSDIENKANVSAICRRRLPVIMCKLRMSQVVSEATRLIEQGHVRVGPHVITDPAY 148
Query 164 MVRVDSEKYIEFSLTSPYGGGRPGRVKRKSLRGGDDKDGDD 204
+V E ++ ++ TS ++KR + D D D
Sbjct 149 LVTRSLEDFVTWTDTS--------KIKRTIAKYNDKLDDYD 181
> CE08804
Length=183
Score = 54.7 bits (130), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 39/124 (31%), Positives = 60/124 (48%), Gaps = 6/124 (4%)
Query 61 RVQYAL-----AKIRSAARELLTLEEKDPRRIFQGTALLRRMVRLGLLSEGEQKLDYVLG 115
R YAL AK R A + L E DP R +L + GL+ + L+ +
Sbjct 41 REHYALYNTLAAKSREVADLIKNLSESDPFRSKCTEDMLTKFYAAGLVPTSDT-LERIGK 99
Query 116 LTVQKFLERRLQTRVFKLGLAKSIHHARCLIRQRHIRVGKQIVDIPSFMVRVDSEKYIEF 175
+T F RRL + +G+ +S+ A L+ Q H+R+G ++V P+FMV SE I +
Sbjct 100 VTGASFARRRLPVVMRNIGMCESVKTASDLVEQGHVRIGTKLVTDPAFMVTRSSEDMITW 159
Query 176 SLTS 179
+ S
Sbjct 160 TKAS 163
> ECU05g1320
Length=180
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 34/128 (26%), Positives = 64/128 (50%), Gaps = 2/128 (1%)
Query 48 LGEYGLKNKREVWRVQYALAKIRSAARELLTLEEKDPRRIFQGTALLRRMVRLGLLSEGE 107
+ +YGL+++ E + + K+R A L L+E DP R L+ R+ +G++
Sbjct 32 IAKYGLRDREEYTKYSRIVGKVRKLAEALARLKESDPVRTTITKKLVGRLYSVGVIK--N 89
Query 108 QKLDYVLGLTVQKFLERRLQTRVFKLGLAKSIHHARCLIRQRHIRVGKQIVDIPSFMVRV 167
+KL +TV F ERRL + ++ + S A + H+R+G ++V PS ++
Sbjct 90 RKLVDCTKVTVSSFCERRLPMVMKRIRMVPSFKDATRFVEHGHVRLGSKVVYDPSTIISK 149
Query 168 DSEKYIEF 175
E ++ +
Sbjct 150 AMEDFVSW 157
> At4g12160
Length=106
Score = 52.8 bits (125), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 25/33 (75%), Positives = 28/33 (84%), Gaps = 0/33 (0%)
Query 107 EQKLDYVLGLTVQKFLERRLQTRVFKLGLAKSI 139
+ KLDYVL LTV+ FLERRLQT VFK G+AKSI
Sbjct 20 KNKLDYVLALTVENFLERRLQTIVFKSGMAKSI 52
> At5g15750
Length=194
Score = 52.8 bits (125), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 37/126 (29%), Positives = 59/126 (46%), Gaps = 9/126 (7%)
Query 79 LEEKDPRRIFQGTALLRRMVRLGLLSEGEQKLDYVLGLTVQKFLERRLQTRVFKLGLAKS 138
++ DP RI LL ++ +G++ + L L+V F RRL T + L A+
Sbjct 75 MDPADPFRIQMTDMLLEKLYNMGVIP-TRKSLTLTERLSVSSFCRRRLSTVLVHLKFAEH 133
Query 139 IHHARCLIRQRHIRVGKQIVDIPSFMVRVDSEKYIEFSLTSPYGGGRPGRVKRKSLRGGD 198
A I Q H+RVG + + P+F+V + E +I + +S ++KRK L D
Sbjct 134 HKEAVTYIEQGHVRVGPETITDPAFLVTRNMEDFITWVDSS--------KIKRKVLEYND 185
Query 199 DKDGDD 204
D D
Sbjct 186 TLDDYD 191
> AtCh025
Length=201
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 32/87 (36%), Positives = 45/87 (51%), Gaps = 8/87 (9%)
Query 79 LEEKDPRRIFQGTA--LLRRMVRLGLLSEGEQKLDYVLGLTVQKFLERRLQTRVFKLGLA 136
LEEK R G L + VR+ ++G G + + LE RL +F+LG+A
Sbjct 48 LEEKQKLRFHYGLTEHQLLKYVRIAGKAKGST------GQVLLQLLEMRLDNILFRLGMA 101
Query 137 KSIHHARCLIRQRHIRVGKQIVDIPSF 163
+I AR L+ HI V +IVDIPS+
Sbjct 102 LTIPQARQLVNHGHILVNGRIVDIPSY 128
> 7302772
Length=200
Score = 36.6 bits (83), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 37/178 (20%), Positives = 76/178 (42%), Gaps = 28/178 (15%)
Query 43 QELRLLGEYGLKNKREVWRVQYALAKIRSAARELLTLEEKDPRRIFQGTALLRRMVRLGL 102
+E ++L + ++ + + + +IR A + L+ +P + T LL ++ +G+
Sbjct 28 KENKILRRFHIQKREDYTKYNKLSREIRELAERIAKLDASEPFKTEATTMLLNKLHAMGV 87
Query 103 LSEGEQKLDYVLGLTVQKFLERRLQTRVFK------LGLAKSIHH-------------AR 143
S + L+ ++ F RRL + K + + S +H A
Sbjct 88 -SNDQLTLETAAKISASHFCRRRLPVIMVKREYHPLISTSFSYNHFPLAVRMSEHLKAAT 146
Query 144 CLIRQRHIRVGKQIVDIPSFMVRVDSEKYIEFSLTSPYGGGRPGRVKRKSLRGGDDKD 201
LI H+RVG +++ P+F+V + E ++ + S ++K LR D +D
Sbjct 147 DLIEHGHVRVGPEMIKDPAFLVSRNLEDFVTWVDGS--------KIKEHVLRYNDMRD 196
> CE08539
Length=676
Score = 29.3 bits (64), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 21/56 (37%), Positives = 29/56 (51%), Gaps = 10/56 (17%)
Query 154 GKQIVDIPSF----MVRVDSEKY-----IEFSLTSPYGGGRPGRVKRKSLRGGDDK 200
G+ I+ P F V+V+ EK IEF LT P+ GG+P R++ R G K
Sbjct 503 GRSIMSTPWFEFGGKVKVECEKTGYRADIEF-LTKPFFGGKPHRIQGSICRDGAKK 557
Lambda K H
0.322 0.140 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3658712052
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40