bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0754_orf1
Length=356
Score E
Sequences producing significant alignments: (Bits) Value
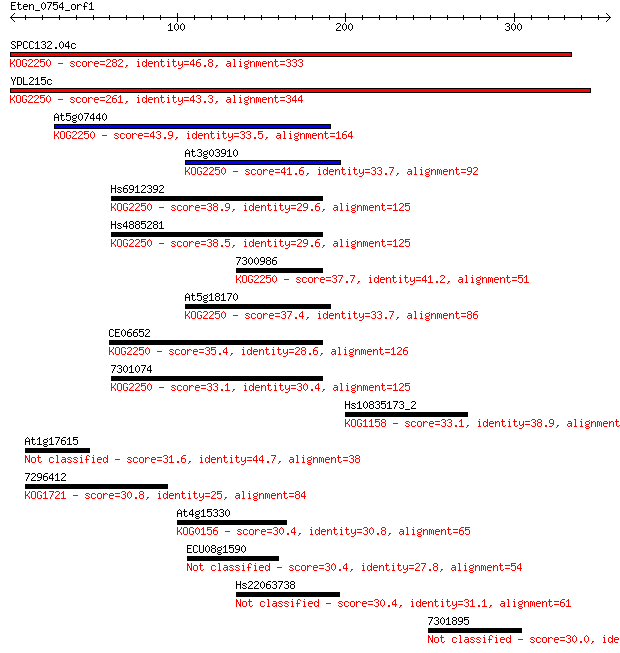
SPCC132.04c 282 1e-75
YDL215c 261 1e-69
At5g07440 43.9 6e-04
At3g03910 41.6 0.003
Hs6912392 38.9 0.016
Hs4885281 38.5 0.023
7300986 37.7 0.041
At5g18170 37.4 0.056
CE06652 35.4 0.18
7301074 33.1 0.95
Hs10835173_2 33.1 0.98
At1g17615 31.6 3.0
7296412 30.8 5.2
At4g15330 30.4 5.5
ECU08g1590 30.4 5.7
Hs22063738 30.4 6.1
7301895 30.0 7.1
> SPCC132.04c
Length=1106
Score = 282 bits (721), Expect = 1e-75, Method: Compositional matrix adjust.
Identities = 156/333 (46%), Positives = 205/333 (61%), Gaps = 12/333 (3%)
Query 1 QTGGPDGDLGSNALLQTKSKTLAVVDGAGVVYDPEGLNAEELRRLARLRFKGEKTSSMLF 60
QTGGPDGDLGSN + + K +AV+DG+GV+YDP GL+ EL RLA E+ + F
Sbjct 776 QTGGPDGDLGSNEIKLSNEKYIAVIDGSGVLYDPAGLDRTELLRLA-----DERKTIDHF 830
Query 61 DSSLLSPKGFKVPQDARDITLPDGTYVASGIEFRNNFHLSKFAVCDLFNPCGGRPASVNP 120
D+ LSP+G++V ++ LP+G V +G FRN HL A D F PCGGRP ++N
Sbjct 831 DAGKLSPEGYRVLVKDTNLKLPNGEIVRNGTIFRNTAHLRYKA--DTFVPCGGRPNAINI 888
Query 121 RNVEQLFEQQTGLPKFKFIVEGANVFITDEARRELENRGVILFKDASTNKGGVTSSSMEV 180
NVEQL + G P FK++VEGAN+FIT +A+ LE GVI+ +DAS NKGGVTSSS+EV
Sbjct 889 NNVEQLIDDH-GRPAFKYLVEGANLFITQDAKSVLEKAGVIVIRDASANKGGVTSSSLEV 947
Query 181 LAALSLTEEEFDRHMRVKDPNNPPKFYKDYVEAILQRIQENARLEFEAIWNEAERTGMHK 240
LA+LS + F +M V D P FY DYV + + IQ NA LEFEAIW +
Sbjct 948 LASLSFDDASFKENMCVHD-GKVPTFYADYVNEVKRIIQRNANLEFEAIWKGHSENKIPY 1006
Query 241 CDLTDILSQKINQLKTDMAGSSALWKDEALVNFVLTKSIPDVLVPGLISVETFRQRVPET 300
L++ LS +I +L D+ LW D N VL SIP L I +E +R+PE+
Sbjct 1007 TSLSNHLSTEIVKLDHDIYNYEKLWADVGFRNAVLRASIPKTL-QAKIGLEKMLERIPES 1065
Query 301 YQRAIFTTFLASRFLYKQPFTAEASAFAFIAYL 333
Y RAIF+T+LASRF+Y+ + FAF Y+
Sbjct 1066 YLRAIFSTYLASRFVYQH--VVSSDPFAFFDYI 1096
> YDL215c
Length=1092
Score = 261 bits (668), Expect = 1e-69, Method: Compositional matrix adjust.
Identities = 149/352 (42%), Positives = 214/352 (60%), Gaps = 17/352 (4%)
Query 1 QTGGPDGDLGSNALLQTKSK--TLAVVDGAGVVYDPEGLNAEELRRLARLRFKGEKTSSM 58
QTGGPDGDLGSN +L + LA++DG+GV+ DP+GL+ +EL RLA E+
Sbjct 750 QTGGPDGDLGSNEILLSSPNECYLAILDGSGVLCDPKGLDKDELCRLAH-----ERKMIS 804
Query 59 LFDSSLLSPKGFKVPQDARDITLPDGTYVASGIEFRNNFHLSKFAVCD---LFNPCGGRP 115
FD+S LS GF V DA DI LP+GT VA+G FRN FH F D +F PCGGRP
Sbjct 805 DFDTSKLSNNGFFVSVDAMDIMLPNGTIVANGTTFRNTFHTQIFKFVDHVDIFVPCGGRP 864
Query 116 ASVNPRNVEQLFEQQTGLPKFKFIVEGANVFITDEARRELENRGVILFKDASTNKGGVTS 175
S+ N+ +++TG K +IVEGAN+FIT A+ LE G ILFKDAS NKGGVTS
Sbjct 865 NSITLNNLHYFVDEKTGKCKIPYIVEGANLFITQPAKNALEEHGCILFKDASANKGGVTS 924
Query 176 SSMEVLAALSLTEEEFDRHMRVKDPNNP-PKFYKDYVEAILQRIQENARLEFEAIWNEAE 234
SSMEVLA+L+L + +F H + D + YK YV + RIQ+NA LEF +WN +
Sbjct 925 SSMEVLASLALNDNDF-VHKFIGDVSGERSALYKSYVVEVQSRIQKNAELEFGQLWNLNQ 983
Query 235 RTGMHKCDLTDILSQKINQLKTDMAGSSALW-KDEALVNFVLT-KSIPDVLVPGLISVET 292
G H ++++ LS IN+L D+ S LW D L N++L K IP +L+ + ++
Sbjct 984 LNGTHISEISNQLSFTINKLNDDLVASQELWLNDLKLRNYLLLDKIIPKILI-DVAGPQS 1042
Query 293 FRQRVPETYQRAIFTTFLASRFLYKQPFTAEASAFAFIAYLDEIRSQMAAAA 344
+ +PE+Y + + +++L+S F+Y+ + + F+ ++ ++ + A+A
Sbjct 1043 VLENIPESYLKVLLSSYLSSTFVYQNGI--DVNIGKFLEFIGGLKREAEASA 1092
> At5g07440
Length=411
Score = 43.9 bits (102), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 55/187 (29%), Positives = 80/187 (42%), Gaps = 35/187 (18%)
Query 27 GAGVVYDPEGLNAEELRRLARLRFKGEKTSSM-LFDSSLLSPKGFKVPQDARDITL---- 81
G GVV+ E L AE + + L F + ++ + + L+ KG KV DIT
Sbjct 186 GRGVVFATEALLAEYGKSIQGLTFVIQGFGNVGTWAAKLIHEKGGKVVA-VSDITGAIRN 244
Query 82 PDGTYVASGIEFR------NNFHLSK--------FAVCDLFNPC--GGRPASVNPRNVEQ 125
P+G + + I+ + N+F+ CD+ PC GG
Sbjct 245 PEGIDINALIKHKDATGSLNDFNGGDAMNSDELLIHECDVLIPCALGG-----------V 293
Query 126 LFEQQTGLPKFKFIVEGANVFITDEARRELENRGVILFKDASTNKGGVTSSSMEVLAALS 185
L ++ G K KFIVE AN +A L +GVI+ D N GGVT S E + +
Sbjct 294 LNKENAGDVKAKFIVEAANHPTDPDADEILSKKGVIILPDIYANAGGVTVSYFEWVQNIQ 353
Query 186 --LTEEE 190
+ EEE
Sbjct 354 GFMWEEE 360
> At3g03910
Length=411
Score = 41.6 bits (96), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 31/97 (31%), Positives = 45/97 (46%), Gaps = 16/97 (16%)
Query 105 CDLFNPC--GGRPASVNPRNVEQLFEQQTGLPKFKFIVEGANVFITDEARRELENRGVIL 162
CD+ P GG +N N ++ K KFI+EGAN EA L+ +GV++
Sbjct 282 CDILVPAALGG---VINRENANEI--------KAKFIIEGANHPTDPEADEILKKKGVMI 330
Query 163 FKDASTNKGGVTSSSMEV---LAALSLTEEEFDRHMR 196
D N GGVT S E + EE+ +R ++
Sbjct 331 LPDIYANSGGVTVSYFEWVQNIQGFMWDEEKVNRELK 367
> Hs6912392
Length=558
Score = 38.9 bits (89), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 37/126 (29%), Positives = 52/126 (41%), Gaps = 13/126 (10%)
Query 61 DSSLLSPKGFKVPQDARDITLPDGTYVASGIEFRNNFHLSKFAV-CDLFNPCGGRPASVN 119
D S+ +P G P++ D L G+ + G + S V CD+ P
Sbjct 334 DGSIWNPDGID-PKELEDFKLQHGSIL--GFPKAKPYEGSILEVDCDILIPAATE----- 385
Query 120 PRNVEQLFEQQTGLPKFKFIVEGANVFITDEARRELENRGVILFKDASTNKGGVTSSSME 179
+QL + K K I EGAN T EA + R +++ D N GGVT S E
Sbjct 386 ----KQLTKSNAPRVKAKIIAEGANGPTTPEADKIFLERNILVIPDLYLNAGGVTVSYFE 441
Query 180 VLAALS 185
L L+
Sbjct 442 WLKNLN 447
> Hs4885281
Length=558
Score = 38.5 bits (88), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 37/126 (29%), Positives = 52/126 (41%), Gaps = 13/126 (10%)
Query 61 DSSLLSPKGFKVPQDARDITLPDGTYVASGIEFRNNFHLSKF-AVCDLFNPCGGRPASVN 119
D S+ +P G P++ D L G+ + G + S A CD+ P
Sbjct 334 DGSIWNPDGID-PKELEDFKLQHGSIL--GFPKAKPYEGSILEADCDILIPAASE----- 385
Query 120 PRNVEQLFEQQTGLPKFKFIVEGANVFITDEARRELENRGVILFKDASTNKGGVTSSSME 179
+QL + K K I EGAN T EA + R +++ D N GGVT S E
Sbjct 386 ----KQLTKSNAPRVKAKIIAEGANGPTTPEADKIFLERNIMVIPDLYLNAGGVTVSYFE 441
Query 180 VLAALS 185
L L+
Sbjct 442 WLKNLN 447
> 7300986
Length=520
Score = 37.7 bits (86), Expect = 0.041, Method: Compositional matrix adjust.
Identities = 21/51 (41%), Positives = 28/51 (54%), Gaps = 0/51 (0%)
Query 135 KFKFIVEGANVFITDEARRELENRGVILFKDASTNKGGVTSSSMEVLAALS 185
K K I+EGAN T + L ++GV+L D N GGVT S E L ++
Sbjct 361 KAKLILEGANGPTTPSGEKILLDKGVLLVPDLYCNAGGVTVSYFEYLKNIN 411
> At5g18170
Length=411
Score = 37.4 bits (85), Expect = 0.056, Method: Compositional matrix adjust.
Identities = 29/90 (32%), Positives = 41/90 (45%), Gaps = 15/90 (16%)
Query 105 CDLFNPC--GGRPASVNPRNVEQLFEQQTGLPKFKFIVEGANVFITDEARRELENRGVIL 162
CD+ P GG +N N ++ K KFI+E AN +A L +GV++
Sbjct 282 CDILVPAALGG---VINRENANEI--------KAKFIIEAANHPTDPDADEILSKKGVVI 330
Query 163 FKDASTNKGGVTSSSMEVLAALS--LTEEE 190
D N GGVT S E + + + EEE
Sbjct 331 LPDIYANSGGVTVSYFEWVQNIQGFMWEEE 360
> CE06652
Length=536
Score = 35.4 bits (80), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 36/128 (28%), Positives = 55/128 (42%), Gaps = 12/128 (9%)
Query 60 FDSSLLSPKGFKVPQDARDITLPDGTYVA-SGIEFRNNFHLSKFAVCDLFNPCGGRPASV 118
+D ++ +P G P++ D +GT G + + F + CD+F P S+
Sbjct 311 YDCAVYNPDGIH-PKELEDWKDANGTIKNFPGAKNFDPFTELMYEKCDIFVPAACE-KSI 368
Query 119 NPRNVEQLFEQQTGLPKFKFIVEGANVFITDEARRELENRG-VILFKDASTNKGGVTSSS 177
+ N ++ + K I E AN T A R L RG ++ D N GGVT S
Sbjct 369 HKENASRI--------QAKIIAEAANGPTTPAADRILLARGDCLIIPDMYVNSGGVTVSY 420
Query 178 MEVLAALS 185
E L L+
Sbjct 421 FEWLKNLN 428
> 7301074
Length=590
Score = 33.1 bits (74), Expect = 0.95, Method: Compositional matrix adjust.
Identities = 38/130 (29%), Positives = 53/130 (40%), Gaps = 20/130 (15%)
Query 61 DSSLLSPKGFKVPQDARDITLPDGTYV----ASGIEFRNNFHLSKFAVCDLFNPCGGRPA 116
D +L +P+G P+ D GT V A E N F CD+F P
Sbjct 327 DGTLYNPEGID-PKLLEDYKNEHGTIVGYQNAKPYEGENLM----FEKCDIFIPAA---- 377
Query 117 SVNPRNVEQLF-EQQTGLPKFKFIVEGANVFITDEARRELENRGVILFKDASTNKGGVTS 175
VE++ + + K I E AN T A + L +R +++ D N GGVT
Sbjct 378 ------VEKVITSENANRIQAKIIAEAANGPTTPAADKILIDRNILVIPDLYINAGGVTV 431
Query 176 SSMEVLAALS 185
S E L L+
Sbjct 432 SFFEWLKNLN 441
> Hs10835173_2
Length=1157
Score = 33.1 bits (74), Expect = 0.98, Method: Compositional matrix adjust.
Identities = 28/87 (32%), Positives = 41/87 (47%), Gaps = 15/87 (17%)
Query 200 PNNPPKF--YKDY------VEAILQRIQENARLEFEAI----WNEAERTGMHK-CDLT-- 244
P PKF +KD + A+ + E LEF A W G+ CD +
Sbjct 273 PIRHPKFEWFKDLGLKWYGLPAVSNMLLEIGGLEFSACPFSGWYMGTEIGVRDYCDNSRY 332
Query 245 DILSQKINQLKTDMAGSSALWKDEALV 271
+IL + ++ DM +S+LWKD+ALV
Sbjct 333 NILEEVAKKMNLDMRKTSSLWKDQALV 359
> At1g17615
Length=380
Score = 31.6 bits (70), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 17/38 (44%), Positives = 22/38 (57%), Gaps = 0/38 (0%)
Query 10 GSNALLQTKSKTLAVVDGAGVVYDPEGLNAEELRRLAR 47
GS L+ T+ K L V G VVY+ E L E+R+L R
Sbjct 318 GSIVLITTQDKQLLVAFGIKVVYEVECLRCFEVRQLFR 355
> 7296412
Length=996
Score = 30.8 bits (68), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 21/95 (22%), Positives = 41/95 (43%), Gaps = 11/95 (11%)
Query 10 GSNALLQTKSKTLAVVDGAGVVYDPEGLNAEELRRLA-----RLRFKGEKTSSMLFDSSL 64
G+ ++Q + ++ GV+ EGL E L+ + + G+ L D+SL
Sbjct 817 GNQIIVQDPEQIQQLLQSVGVLQSGEGLEGETLQMMTDGSGQMVLVHGDNNEQQLIDASL 876
Query 65 LSPKGFKVPQDARD------ITLPDGTYVASGIEF 93
L+ +G + Q +D + DGT + + +
Sbjct 877 LNSEGQLIIQQGQDGEEGAHVISEDGTRIPVSVSY 911
> At4g15330
Length=517
Score = 30.4 bits (67), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 20/65 (30%), Positives = 28/65 (43%), Gaps = 5/65 (7%)
Query 100 SKFAVCDLFNPCGGRPASVNPRNVEQLFEQQTGLPKFKFIVEGANVFITDEARRELENRG 159
S+F L+ C GR SV VE++ E L F++ R+ LE G
Sbjct 183 SRFVNNSLYKMCTGRSFSVENNEVERIMELTADLGAL-----SQKFFVSKMFRKLLEKLG 237
Query 160 VILFK 164
+ LFK
Sbjct 238 ISLFK 242
> ECU08g1590
Length=390
Score = 30.4 bits (67), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 15/54 (27%), Positives = 26/54 (48%), Gaps = 0/54 (0%)
Query 106 DLFNPCGGRPASVNPRNVEQLFEQQTGLPKFKFIVEGANVFITDEARRELENRG 159
D+FN S N+++++ +T K + +G N + DE R+ NRG
Sbjct 285 DVFNELENGNYSSALENLKRMYRVETTTDLGKHLKDGQNTLVLDEIRKSYSNRG 338
> Hs22063738
Length=605
Score = 30.4 bits (67), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 19/61 (31%), Positives = 30/61 (49%), Gaps = 8/61 (13%)
Query 135 KFKFIVEGANVFITDEARRELENRGVILFKDASTNKGGVTSSSMEVLAALSLTEEEFDRH 194
KFK+ V G N+ ++R+ + D NKG + + ++ ALSL EEE +
Sbjct 468 KFKWSVNGWNLIAVQPSKRK--------YVDIFQNKGNKSCPAWKISIALSLEEEESNNK 519
Query 195 M 195
M
Sbjct 520 M 520
> 7301895
Length=212
Score = 30.0 bits (66), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 19/56 (33%), Positives = 32/56 (57%), Gaps = 5/56 (8%)
Query 249 QKINQLKTDMAGSSALWKDEALVNFV-LTKSIPDVLVPGLISVETFRQRVPETYQR 303
Q++ + + + GSS++ A+V F L K IPD+L P ++E F QR + +R
Sbjct 160 QQLRRYEQQLRGSSSV----AMVYFASLDKEIPDILEPEYRAMEDFYQRYRSSLKR 211
Lambda K H
0.317 0.133 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8629504356
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40