bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0781_orf1
Length=192
Score E
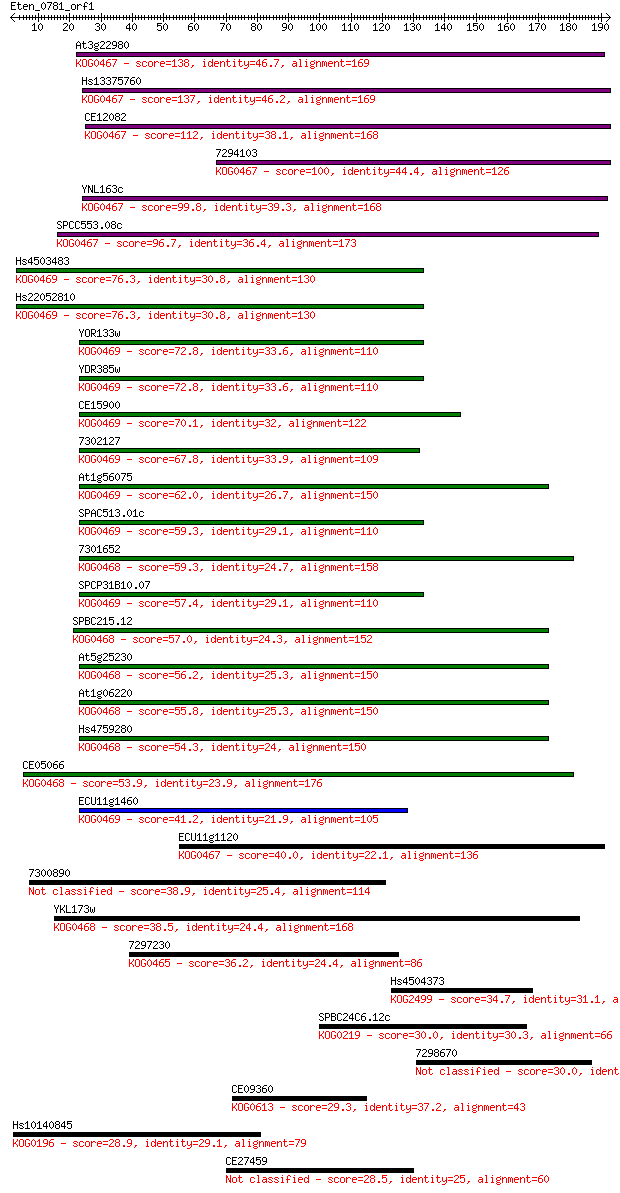
Sequences producing significant alignments: (Bits) Value
At3g22980 138 6e-33
Hs13375760 137 1e-32
CE12082 112 4e-25
7294103 100 2e-21
YNL163c 99.8 3e-21
SPCC553.08c 96.7 3e-20
Hs4503483 76.3 3e-14
Hs22052810 76.3 3e-14
YOR133w 72.8 4e-13
YDR385w 72.8 4e-13
CE15900 70.1 3e-12
7302127 67.8 1e-11
At1g56075 62.0 8e-10
SPAC513.01c 59.3 5e-09
7301652 59.3 5e-09
SPCP31B10.07 57.4 2e-08
SPBC215.12 57.0 2e-08
At5g25230 56.2 4e-08
At1g06220 55.8 4e-08
Hs4759280 54.3 1e-07
CE05066 53.9 2e-07
ECU11g1460 41.2 0.001
ECU11g1120 40.0 0.003
7300890 38.9 0.007
YKL173w 38.5 0.009
7297230 36.2 0.046
Hs4504373 34.7 0.12
SPBC24C6.12c 30.0 3.3
7298670 30.0 3.3
CE09360 29.3 4.6
Hs10140845 28.9 6.2
CE27459 28.5 8.5
> At3g22980
Length=1015
Score = 138 bits (348), Expect = 6e-33, Method: Compositional matrix adjust.
Identities = 79/171 (46%), Positives = 114/171 (66%), Gaps = 9/171 (5%)
Query 22 VGSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEE 81
+ +GQV++A+K+ACRAA+LQ RI EAM L+ +G Y+VL+RRR +I+KEE
Sbjct 850 IFTGQVMTAVKDACRAAVLQTN-PRIVEAMYFCELNTAPEYLGPMYAVLSRRRARILKEE 908
Query 82 VPEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPF--PEACMTE 139
+ EG + F V ++P +E+ G + E+R SG AS + SHWE+L+E+PF P+ TE
Sbjct 909 MQEGSSLFTVHAYVPVSESFGFADELRKGTSGGASALMVLSHWEMLEEDPFFVPK---TE 965
Query 140 QELEDEGEAALSSLATQIHARAIINSIRKSKGLPTEEKVVNDAEKQRTLSR 190
+E+E+ G+ A S L AR +IN++R+ KGL EEKVV A KQRTL+R
Sbjct 966 EEIEEFGDGA-SVLPNT--ARKLINAVRRRKGLHVEEKVVQYATKQRTLAR 1013
> Hs13375760
Length=857
Score = 137 bits (345), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 78/169 (46%), Positives = 107/169 (63%), Gaps = 6/169 (3%)
Query 24 SGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEVP 83
SGQ+I+ MKEACR A LQ R+ AM + V+G+ Y+VL++R G++++EE+
Sbjct 695 SGQLIATMKEACRYA-LQVKPQRLMAAMYTCDIMATGDVLGRVYAVLSKREGRVLQEEMK 753
Query 84 EGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQELE 143
EG F++ LP AE+ G + EIR + SG AS QL FSHWE++ +PF TE+E
Sbjct 754 EGTDMFIIKAVLPVAESFGFADEIRKRTSGLASPQLVFSHWEIIPSDPF-WVPTTEEEYL 812
Query 144 DEGEAALSSLATQIHARAIINSIRKSKGLPTEEKVVNDAEKQRTLSRNK 192
GE A S + AR +N++RK KGL EEK+V AEKQRTLS+NK
Sbjct 813 HFGEKADS----ENQARKYMNAVRKRKGLYVEEKIVEHAEKQRTLSKNK 857
> CE12082
Length=906
Score = 112 bits (281), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 64/180 (35%), Positives = 102/180 (56%), Gaps = 18/180 (10%)
Query 25 GQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKI------- 77
GQ+++A+K +C AA + L R+ AM R T++ + +GK ++VL++R+ K+
Sbjct 733 GQMMTAIKASCSAAAKKLAL-RLVAAMYRCTVTTASQALGKVHAVLSQRKSKVGMENCRT 791
Query 78 -----IKEEVPEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPF 132
+ E++ E F V +P E+ ++R SG AS QLQFSHW+V+ E+P+
Sbjct 792 FSNIVLSEDINEATNLFEVVSLMPVVESFSFCDQLRKFTSGMASAQLQFSHWQVIDEDPY 851
Query 133 PEACMTEQELEDEGEAALSSLATQIHARAIINSIRKSKGLPTEEKVVNDAEKQRTLSRNK 192
T LE+ E L + HAR ++++R+ KGLPTE+ +V AEKQR L +NK
Sbjct 852 ----WTPSTLEEIEEFGLKGDSPN-HARGYMDAVRRRKGLPTEDLIVESAEKQRNLKKNK 906
> 7294103
Length=122
Score = 100 bits (248), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 56/126 (44%), Positives = 79/126 (62%), Gaps = 5/126 (3%)
Query 67 YSVLARRRGKIIKEEVPEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEV 126
Y+V+ RR GKI+ ++ +G +F V LP E+ +QE+R + SG A QL FSHWEV
Sbjct 2 YAVIGRRHGKILSGDLTQGSGNFAVTCLLPVIESFNFAQEMRKQTSGLACPQLMFSHWEV 61
Query 127 LQEEPFPEACMTEQELEDEGEAALSSLATQIHARAIINSIRKSKGLPTEEKVVNDAEKQR 186
+ +PF TE+EL GE A S+ A+ ++S+R+ KGL +E+VV AEKQR
Sbjct 62 IDIDPFWLPT-TEEELMHFGEKADSA----NRAKVYMDSVRRRKGLFVDEQVVEHAEKQR 116
Query 187 TLSRNK 192
TLS+NK
Sbjct 117 TLSKNK 122
> YNL163c
Length=1110
Score = 99.8 bits (247), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 66/170 (38%), Positives = 96/170 (56%), Gaps = 10/170 (5%)
Query 24 SGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEVP 83
SG++I++ ++A A L RI A+ + V+GK Y+V+ +R GKII EE+
Sbjct 949 SGRLITSTRDAIHEAFLDWS-PRIMWAIYSCDIQTSVDVLGKVYAVILQRHGKIISEEMK 1007
Query 84 EGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPF--PEACMTEQE 141
EG F + +P EA G+S++IR + SG A QL FS +E + +PF P +E
Sbjct 1008 EGTPFFQIEAHVPVVEAFGLSEDIRKRTSGAAQPQLVFSGFECIDLDPFWVPTTEEELEE 1067
Query 142 LEDEGEAALSSLATQIHARAIINSIRKSKGLPTEEKVVNDAEKQRTLSRN 191
L D + + AR +N+IR+ KGL EEKVV +AEKQRTL +N
Sbjct 1068 LGDTAD-------RENIARKHMNAIRRRKGLFIEEKVVENAEKQRTLKKN 1110
> SPCC553.08c
Length=1000
Score = 96.7 bits (239), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 63/173 (36%), Positives = 95/173 (54%), Gaps = 6/173 (3%)
Query 16 LSVHSYVGSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRG 75
L++++ GQVIS +KE+ R L R+ AM + V+G+ Y V+++RRG
Sbjct 832 LTINNPQIPGQVISVVKESIRHGFLGWS-PRLMLAMYSCDVQATSEVLGRVYGVVSKRRG 890
Query 76 KIIKEEVPEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEA 135
++I EE+ EG F+V +P E+ G + EI + SG A QL F +E+L E PF
Sbjct 891 RVIDEEMKEGTPFFIVKALIPVVESFGFAVEILKRTSGAAYPQLIFHGFEMLDENPFWVP 950
Query 136 CMTEQELEDEGEAALSSLATQIHARAIINSIRKSKGLPTEEKVVNDAEKQRTL 188
E+ + A ++A R ++N +RK KGL E+K+V AEKQRTL
Sbjct 951 TTEEELEDLGELADRENIA----KRYMLN-VRKRKGLLVEQKIVEKAEKQRTL 998
> Hs4503483
Length=858
Score = 76.3 bits (186), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 40/133 (30%), Positives = 68/133 (51%), Gaps = 4/133 (3%)
Query 3 ESKLTGKQTTTGPLSVHS---YVGSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCD 59
E + G + +++H+ + G GQ+I + A++L R+ E + + C
Sbjct 694 EENMRGVRFDVHDVTLHADAIHRGGGQIIPTARRCLYASVLT-AQPRLMEPIYLVEIQCP 752
Query 60 QRVIGKAYSVLARRRGKIIKEEVPEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQL 119
++V+G Y VL R+RG + +E G FVV +LP E+ G + ++RS G A Q
Sbjct 753 EQVVGGIYGVLNRKRGHVFEESQVAGTPMFVVKAYLPVNESFGFTADLRSNTGGQAFPQC 812
Query 120 QFSHWEVLQEEPF 132
F HW++L +PF
Sbjct 813 VFDHWQILPGDPF 825
> Hs22052810
Length=846
Score = 76.3 bits (186), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 40/133 (30%), Positives = 68/133 (51%), Gaps = 4/133 (3%)
Query 3 ESKLTGKQTTTGPLSVHS---YVGSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCD 59
E + G + +++H+ + G GQ+I + A++L R+ E + + C
Sbjct 682 EENMRGVRFDVHDVTLHADAIHRGGGQIIPTARRCLYASVLT-AQPRLMEPIYLVEIQCP 740
Query 60 QRVIGKAYSVLARRRGKIIKEEVPEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQL 119
++V+G Y VL R+RG + +E G FVV +LP E+ G + ++RS G A Q
Sbjct 741 EQVVGGIYGVLNRKRGHVFEESQVAGTPMFVVKAYLPVNESFGFTADLRSNTGGQAFPQC 800
Query 120 QFSHWEVLQEEPF 132
F HW++L +PF
Sbjct 801 VFDHWQILPGDPF 813
> YOR133w
Length=842
Score = 72.8 bits (177), Expect = 4e-13, Method: Composition-based stats.
Identities = 37/110 (33%), Positives = 56/110 (50%), Gaps = 1/110 (0%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
G GQ+I M+ A A L +I E + + C ++ +G YSVL ++RG+++ EE
Sbjct 701 GGGQIIPTMRRATYAGFLLAD-PKIQEPVFLVEIQCPEQAVGGIYSVLNKKRGQVVSEEQ 759
Query 83 PEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPF 132
G F V +LP E+ G + E+R G A Q+ F HW L +P
Sbjct 760 RPGTPLFTVKAYLPVNESFGFTGELRQATGGQAFPQMVFDHWSTLGSDPL 809
> YDR385w
Length=842
Score = 72.8 bits (177), Expect = 4e-13, Method: Composition-based stats.
Identities = 37/110 (33%), Positives = 56/110 (50%), Gaps = 1/110 (0%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
G GQ+I M+ A A L +I E + + C ++ +G YSVL ++RG+++ EE
Sbjct 701 GGGQIIPTMRRATYAGFLLAD-PKIQEPVFLVEIQCPEQAVGGIYSVLNKKRGQVVSEEQ 759
Query 83 PEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPF 132
G F V +LP E+ G + E+R G A Q+ F HW L +P
Sbjct 760 RPGTPLFTVKAYLPVNESFGFTGELRQATGGQAFPQMVFDHWSTLGSDPL 809
> CE15900
Length=852
Score = 70.1 bits (170), Expect = 3e-12, Method: Composition-based stats.
Identities = 39/122 (31%), Positives = 58/122 (47%), Gaps = 1/122 (0%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
G GQ+I + A++L R+ E + + C + +G Y VL RRRG + +E
Sbjct 711 GGGQIIPTARRVFYASVLT-AEPRLLEPVYLVEIQCPEAAVGGIYGVLNRRRGHVFEESQ 769
Query 83 PEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQEL 142
G FVV +LP E+ G + ++RS G A Q F HW+VL +P Q +
Sbjct 770 VTGTPMFVVKAYLPVNESFGFTADLRSNTGGQAFPQCVFDHWQVLPGDPLEAGTKPNQIV 829
Query 143 ED 144
D
Sbjct 830 LD 831
> 7302127
Length=832
Score = 67.8 bits (164), Expect = 1e-11, Method: Composition-based stats.
Identities = 37/109 (33%), Positives = 54/109 (49%), Gaps = 1/109 (0%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
G GQ+I + AA + R+ E + + C + +G Y VL RRRG + +E
Sbjct 691 GGGQIIPTTRRCLYAAAIT-AKPRLMEPVYLCEIQCPEVAVGGIYGVLNRRRGHVFEENQ 749
Query 83 PEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEP 131
G FVV +LP E+ G + ++RS G A Q F HW+VL +P
Sbjct 750 VVGTPMFVVKAYLPVNESFGFTADLRSNTGGQAFPQCVFDHWQVLPGDP 798
> At1g56075
Length=855
Score = 62.0 bits (149), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 40/150 (26%), Positives = 64/150 (42%), Gaps = 22/150 (14%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
G GQVI + A+ + R+ E + + + +G YSVL ++RG + +E
Sbjct 714 GGGQVIPTARRVIYASQIT-AKPRLLEPVYMVEIQAPEGALGGIYSVLNQKRGHVFEEMQ 772
Query 83 PEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQEL 142
G + + +LP E+ G S ++R+ SG A Q F HWE++ +P
Sbjct 773 RPGTPLYNIKAYLPVVESFGFSSQLRAATSGQAFPQCVFDHWEMMSSDP----------- 821
Query 143 EDEGEAALSSLATQIHARAIINSIRKSKGL 172
L A ++ IRK KGL
Sbjct 822 ----------LEPGTQASVLVADIRKRKGL 841
> SPAC513.01c
Length=812
Score = 59.3 bits (142), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 32/110 (29%), Positives = 51/110 (46%), Gaps = 1/110 (0%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
G GQ+I + A+ L I E + + + +G YSVL ++RG + EE
Sbjct 671 GGGQIIPTARRVVYASTLLASPI-IQEPVFLVEIQVSENAMGGIYSVLNKKRGHVFSEEQ 729
Query 83 PEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPF 132
G + + +LP E+ G + E+R +G A QL F HW + +P
Sbjct 730 RVGTPLYNIKAYLPVNESFGFTGELRQATAGQAFPQLVFDHWSPMSGDPL 779
> 7301652
Length=975
Score = 59.3 bits (142), Expect = 5e-09, Method: Composition-based stats.
Identities = 39/158 (24%), Positives = 72/158 (45%), Gaps = 11/158 (6%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
G GQ+I + +A L R+ E L + + Y+VLARRRG + ++
Sbjct 807 GGGQIIPTARRVAYSAFLM-ATPRLMEPYLFVEVQAPADCVSAVYTVLARRRGHVTQDAP 865
Query 83 PEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQEL 142
G + + F+P ++ G ++R+ G A F HW+++ +P ++ + + L
Sbjct 866 VSGSPIYTIKAFIPAIDSFGFETDLRTHTQGQAFCLSVFHHWQIVPGDPLDKSIII-RPL 924
Query 143 EDEGEAALSSLATQIHARAIINSIRKSKGLPTEEKVVN 180
E + + L AR + R+ KGL +E+ +N
Sbjct 925 EPQQASHL--------AREFMIKTRRRKGL-SEDVSIN 953
> SPCP31B10.07
Length=842
Score = 57.4 bits (137), Expect = 2e-08, Method: Composition-based stats.
Identities = 32/110 (29%), Positives = 51/110 (46%), Gaps = 1/110 (0%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
G GQ+I + A+ L I E + + + +G YSVL ++RG + EE
Sbjct 701 GGGQIIPTARRVVYASTLLASPI-IQEPVFLVEIQVSENAMGGIYSVLNKKRGHVFSEEQ 759
Query 83 PEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPF 132
G + + +LP E+ G + E+R +G A QL F HW + +P
Sbjct 760 RVGTPLYNIKAYLPVNESFGFTGELRQATAGQAFPQLVFDHWSPMSGDPL 809
> SPBC215.12
Length=983
Score = 57.0 bits (136), Expect = 2e-08, Method: Composition-based stats.
Identities = 37/152 (24%), Positives = 68/152 (44%), Gaps = 10/152 (6%)
Query 21 YVGSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKE 80
Y G GQ+I + C ++ L R+ E + + + Y +L RRRG ++++
Sbjct 815 YRGGGQIIPTARRVCYSSFLTAS-PRLMEPVYMVEVHAPADSLPIIYDLLTRRRGHVLQD 873
Query 81 EVPEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQ 140
G ++V +P ++ G ++R G A Q+ F HW+V+ +P ++ + +
Sbjct 874 IPRPGSPLYLVRALIPVIDSCGFETDLRVHTQGQAMCQMVFDHWQVVPGDPLDKS-IKPK 932
Query 141 ELEDEGEAALSSLATQIHARAIINSIRKSKGL 172
LE + L AR + R+ KGL
Sbjct 933 PLEPARGSDL--------ARDFLIKTRRRKGL 956
> At5g25230
Length=973
Score = 56.2 bits (134), Expect = 4e-08, Method: Composition-based stats.
Identities = 38/150 (25%), Positives = 62/150 (41%), Gaps = 10/150 (6%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
GSGQ+I + +A L R+ E + + + Y+VL+RRRG + +
Sbjct 804 GSGQMIPTARRVAYSAFLM-ATPRLMEPVYYVEIQTPIDCVTAIYTVLSRRRGYVTSDVP 862
Query 83 PEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQEL 142
G +++V FLP E+ G ++R G A F HW ++ +P +A
Sbjct 863 QPGTPAYIVKAFLPVIESFGFETDLRYHTQGQAFCLSVFDHWAIVPGDPLDKAIQLR--- 919
Query 143 EDEGEAALSSLATQIHARAIINSIRKSKGL 172
L Q AR + R+ KG+
Sbjct 920 ------PLEPAPIQHLAREFMVKTRRRKGM 943
> At1g06220
Length=987
Score = 55.8 bits (133), Expect = 4e-08, Method: Composition-based stats.
Identities = 38/150 (25%), Positives = 62/150 (41%), Gaps = 10/150 (6%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
GSGQ+I + +A L R+ E + + + Y+VL+RRRG + +
Sbjct 818 GSGQMIPTARRVAYSAFLM-ATPRLMEPVYYVEIQTPIDCVTAIYTVLSRRRGHVTSDVP 876
Query 83 PEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQEL 142
G +++V FLP E+ G ++R G A F HW ++ +P +A
Sbjct 877 QPGTPAYIVKAFLPVIESFGFETDLRYHTQGQAFCLSVFDHWAIVPGDPLDKAIQLR--- 933
Query 143 EDEGEAALSSLATQIHARAIINSIRKSKGL 172
L Q AR + R+ KG+
Sbjct 934 ------PLEPAPIQHLAREFMVKTRRRKGM 957
> Hs4759280
Length=972
Score = 54.3 bits (129), Expect = 1e-07, Method: Composition-based stats.
Identities = 36/150 (24%), Positives = 65/150 (43%), Gaps = 10/150 (6%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
G GQ+I + +A L R+ E + + Y+VLARRRG + ++
Sbjct 804 GGGQIIPTARRVVYSAFLM-ATPRLMEPYYFVEVQAPADCVSAVYTVLARRRGHVTQDAP 862
Query 83 PEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQEL 142
G + + F+P ++ G ++R+ G A F HW+++ +P ++ + + L
Sbjct 863 IPGSPLYTIKAFIPAIDSFGFETDLRTHTQGQAFSLSVFHHWQIVPGDPLDKSIVI-RPL 921
Query 143 EDEGEAALSSLATQIHARAIINSIRKSKGL 172
E + L AR + R+ KGL
Sbjct 922 EPQPAPHL--------AREFMIKTRRRKGL 943
> CE05066
Length=974
Score = 53.9 bits (128), Expect = 2e-07, Method: Composition-based stats.
Identities = 42/176 (23%), Positives = 72/176 (40%), Gaps = 15/176 (8%)
Query 5 KLTGKQTTTGPLSVHSYVGSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIG 64
KL T PL Y G GQ+I + +A L R+ E + +
Sbjct 794 KLLDAAIATEPL----YRGGGQMIPTARRCAYSAFLM-ATPRLMEPYYTVEVVAPADCVA 848
Query 65 KAYSVLARRRGKIIKEEVPEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHW 124
Y+VLA+RRG + + G + + ++P ++ G ++R G A F HW
Sbjct 849 AVYTVLAKRRGHVTTDAPMPGSPMYTISAYIPVMDSFGFETDLRIHTQGQAFCMSAFHHW 908
Query 125 EVLQEEPFPEACMTEQELEDEGEAALSSLATQIHARAIINSIRKSKGLPTEEKVVN 180
+++ +P ++ + + L T AR + R+ KGL +E+ VN
Sbjct 909 QLVPGDPLDKSIVIK---------TLDVQPTPHLAREFMIKTRRRKGL-SEDVSVN 954
> ECU11g1460
Length=850
Score = 41.2 bits (95), Expect = 0.001, Method: Composition-based stats.
Identities = 23/105 (21%), Positives = 50/105 (47%), Gaps = 1/105 (0%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
G Q++ +K C+ +L G + E + ++ G ++L +RG +
Sbjct 709 GINQLLQPVKNLCKGLLLAAGPI-LYEPIYEVEITTPNDYSGAVTTILLSKRGTAEDFKT 767
Query 83 PEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVL 127
G + ++ G LP E+ +++++S + G A ++FSH+ +L
Sbjct 768 LPGNDTTMITGTLPVKESFTFNEDLKSGSRGKAGASMRFSHYSIL 812
> ECU11g1120
Length=678
Score = 40.0 bits (92), Expect = 0.003, Method: Composition-based stats.
Identities = 30/136 (22%), Positives = 55/136 (40%), Gaps = 29/136 (21%)
Query 55 TLSCDQRVIGKAYSVLARRRGKIIKEEVPEGQTSFVVFGFLPTAEAPGISQEIRSKASGH 114
++S + IG+ Y L++ +++E E +V+ +P PG ++IR G
Sbjct 572 SISIQKEFIGETYRCLSKYSYTVLQENYEEKTGFYVLTVLIPQFLYPGFLEDIRVGTKGT 631
Query 115 ASLQLQFSHWEVLQEEPFPEACMTEQELEDEGEAALSSLATQIHARAIINSIRKSKGLPT 174
A L + ++++ + ++ IRK KGL
Sbjct 632 AYLLVGDVGYKLM-----------------------------LDFSKYVDGIRKKKGLFV 662
Query 175 EEKVVNDAEKQRTLSR 190
EEK+V + +QRT R
Sbjct 663 EEKIVEEPSRQRTYKR 678
> 7300890
Length=1093
Score = 38.9 bits (89), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 29/118 (24%), Positives = 54/118 (45%), Gaps = 5/118 (4%)
Query 7 TGKQTTTGPLSVHS-YVGSG---QVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRV 62
G Q + +H+ +G G + A C +L R+ E ++ +
Sbjct 553 VGGQVVETQIRLHNATIGRGTADSFVMATAAQCVQKLLSTSGTRLLEPIMALQIVAPSER 612
Query 63 IGKAYSVLARRRGKIIKEEVPEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQ 120
I + L+RRR +I + +P+G+ + ++ P AE G S +R+ +SG AS+ +Q
Sbjct 613 ISGIMADLSRRRA-LINDVLPKGERNKMILVNAPLAELSGYSSALRTISSGTASMTMQ 669
> YKL173w
Length=1008
Score = 38.5 bits (88), Expect = 0.009, Method: Composition-based stats.
Identities = 41/176 (23%), Positives = 74/176 (42%), Gaps = 23/176 (13%)
Query 15 PLSVHSYVGSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRR 74
P V+ V Q+I MK+AC +L + + E + ++ ++ ++ +RR
Sbjct 826 PSDVNIDVMKSQIIPLMKKACYVGLLT-AIPILLEPIYEVDITVHAPLLPIVEELMKKRR 884
Query 75 GKIIKEEVPEGQTSFV-VFGFLPTAEAPGISQEIRSKASGHASLQLQFSH--W-----EV 126
G I + + T + V G +P E+ G ++R +G QL F H W +V
Sbjct 885 GSRIYKTIKVAGTPLLEVRGQVPVIESAGFETDLRLSTNGLGMCQLYFWHKIWRKVPGDV 944
Query 127 LQEEPFPEACMTEQELEDEGEAALSSLATQIHARAIINSIRKSKGLPTEEKVVNDA 182
L ++ F + A ++SL +R + R+ KG+ T + ND
Sbjct 945 LDKDAF---------IPKLKPAPINSL-----SRDFVMKTRRRKGISTGGFMSNDG 986
> 7297230
Length=729
Score = 36.2 bits (82), Expect = 0.046, Method: Compositional matrix adjust.
Identities = 21/86 (24%), Positives = 39/86 (45%), Gaps = 2/86 (2%)
Query 39 MLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEVPEGQTSFVVFGFLPTA 98
+ Q G +I E ++ ++ + G L++R G I E EG F V+ +P
Sbjct 615 VFQNGSWQILEPIMLVEVTAPEEFQGAVMGHLSKRHGIITGTEGTEGW--FTVYAEVPLN 672
Query 99 EAPGISQEIRSKASGHASLQLQFSHW 124
+ G + E+RS G +++S +
Sbjct 673 DMFGYAGELRSSTQGKGEFTMEYSRY 698
> Hs4504373
Length=556
Score = 34.7 bits (78), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 14/45 (31%), Positives = 24/45 (53%), Gaps = 0/45 (0%)
Query 123 HWEVLQEEPFPEACMTEQELEDEGEAALSSLATQIHARAIINSIR 167
HW ++ ++ FP +T EL ++G +LS + T R +I R
Sbjct 235 HWHIVDDQSFPYQSITFPELSNKGSYSLSHVYTPNDVRMVIEYAR 279
> SPBC24C6.12c
Length=434
Score = 30.0 bits (66), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 20/66 (30%), Positives = 35/66 (53%), Gaps = 5/66 (7%)
Query 100 APGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQELEDEGEAALSSLATQIHA 159
A G +QE++SK G A ++ W + PE MT+Q++ D+ + + S A QI++
Sbjct 365 AQGDTQEVKSKKEGMAIVRDIMRQW---RSNVKPE--MTQQQMMDQFQKVIGSFAQQINS 419
Query 160 RAIINS 165
+ S
Sbjct 420 NNWLQS 425
> 7298670
Length=617
Score = 30.0 bits (66), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 18/57 (31%), Positives = 34/57 (59%), Gaps = 2/57 (3%)
Query 131 PFPEACMTEQELEDEGEAALS-SLATQIHARAIINSIRKSKGLPTEEKVVNDAEKQR 186
P+ ++ M +Q + G+ LS +LA +H R+++ + +GLP + +V D EK+R
Sbjct 283 PWNKSFMDKQTWMERGDDRLSVTLAEIVHIRSVMTKA-ELEGLPMDVRVKEDVEKRR 338
> CE09360
Length=2783
Score = 29.3 bits (64), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 16/44 (36%), Positives = 23/44 (52%), Gaps = 7/44 (15%)
Query 72 RRRGKIIKEEVPEGQTSFVVFGFLPTAEA-PGISQEIRSKASGH 114
R + + +E P VF F PT + PG+S E+R+K GH
Sbjct 2564 RNNERSVADEAPR------VFDFEPTTRSDPGVSVELRAKVIGH 2601
> Hs10140845
Length=1005
Score = 28.9 bits (63), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 23/79 (29%), Positives = 35/79 (44%), Gaps = 4/79 (5%)
Query 2 RESKLTGKQTTTGPLSVHSYVGSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQR 61
R KL + + GPLS + + Q I A C A + R + C AM+R + +
Sbjct 162 RRLKLNTEVRSVGPLSKRGFYLAFQDIGA----CLAILSLRIYYKKCPAMVRNLAAFSEA 217
Query 62 VIGKAYSVLARRRGKIIKE 80
V G S L RG+ ++
Sbjct 218 VTGADSSSLVEVRGQCVRH 236
> CE27459
Length=689
Score = 28.5 bits (62), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 15/60 (25%), Positives = 27/60 (45%), Gaps = 0/60 (0%)
Query 70 LARRRGKIIKEEVPEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQE 129
L RRR + E + LP +E +S+ +R+ SG + +QF ++ + E
Sbjct 616 LLRRRAHFEHSDATESTEIRRICAILPLSETENLSKTVRTLTSGFGDISVQFRGYQQVTE 675
Lambda K H
0.315 0.128 0.358
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3238438140
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40