bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0838_orf3
Length=208
Score E
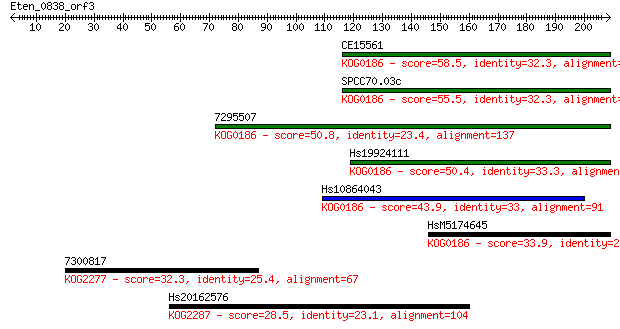
Sequences producing significant alignments: (Bits) Value
CE15561 58.5 8e-09
SPCC70.03c 55.5 7e-08
7295507 50.8 2e-06
Hs19924111 50.4 2e-06
Hs10864043 43.9 2e-04
HsM5174645 33.9 0.21
7300817 32.3 0.73
Hs20162576 28.5 8.9
> CE15561
Length=564
Score = 58.5 bits (140), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 30/93 (32%), Positives = 50/93 (53%), Gaps = 0/93 (0%)
Query 116 SMSDLLRSLFVFSLCRHHWLVDNWESLLNVSCRIFGSRAVFAFIRFSFFKVFCGGEALEE 175
S ++L+R+L V LC LV+ + +LN R+ G ++ +FF F GE EE
Sbjct 20 SNTELVRALVVLRLCGIQTLVNQNQIILNTMRRVLGKNLFKKTLKNTFFGHFVAGETEEE 79
Query 176 VVQTLDKLKQRNLGAILDYAAEQQLPARAAARK 208
V ++KL+ + +ILDY+ E + ++ A K
Sbjct 80 VRHVVEKLRNYGVKSILDYSVEADITSQEATDK 112
> SPCC70.03c
Length=492
Score = 55.5 bits (132), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 30/94 (31%), Positives = 51/94 (54%), Gaps = 2/94 (2%)
Query 116 SMSDLLRSLFVFSLCRHHWLVDNWESLLNVSCRIFGSRAVFA-FIRFSFFKVFCGGEALE 174
S+ +LRS FV+ +C WLV +++ C +F ++ F R++F+K FCGGE +
Sbjct 55 SLFSVLRSAFVYEICSRAWLVKLSLGAMSL-CDVFHLSFLYNPFCRYTFYKHFCGGETPQ 113
Query 175 EVVQTLDKLKQRNLGAILDYAAEQQLPARAAARK 208
V+ T+D L+ + + L+Y+ E L K
Sbjct 114 AVMATMDTLQAAGITSCLNYSREVDLDGDMDVNK 147
> 7295507
Length=671
Score = 50.8 bits (120), Expect = 2e-06, Method: Composition-based stats.
Identities = 32/139 (23%), Positives = 65/139 (46%), Gaps = 11/139 (7%)
Query 72 QEESHQPPSPAPRAPPNQRQQQKQQLQQLVLTGDAAGAAALRLYSMSDLLRSLFVFSLCR 131
++ES Q +P+P P QR + AA + + +L+R+ V+ +C
Sbjct 94 KQESSQEKNPSPAGSP-QRDPLDVSFNDPI--------AAFKSKTTGELIRAYLVYMICS 144
Query 132 HHWLVDN--WESLLNVSCRIFGSRAVFAFIRFSFFKVFCGGEALEEVVQTLDKLKQRNLG 189
LV++ + L+ ++ + G + ++ SF+ F GE +V L++L+ +
Sbjct 145 SEKLVEHNMTKQLMKLARNLLGQKLFVLLMKSSFYGHFVAGENRHTIVPALERLRSFGVK 204
Query 190 AILDYAAEQQLPARAAARK 208
ILDY+ E+ + A ++
Sbjct 205 PILDYSVEEDITQEEAEKR 223
> Hs19924111
Length=600
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 30/90 (33%), Positives = 42/90 (46%), Gaps = 0/90 (0%)
Query 119 DLLRSLFVFSLCRHHWLVDNWESLLNVSCRIFGSRAVFAFIRFSFFKVFCGGEALEEVVQ 178
+L RSL V LC L+ E LL VS ++ G R ++ +F+ F GE E +
Sbjct 69 ELARSLLVLRLCAWPALLARHEQLLYVSRKLLGQRLFNKLMKMTFYGHFVAGEDQESIQP 128
Query 179 TLDKLKQRNLGAILDYAAEQQLPARAAARK 208
L + + AILDY E+ L A K
Sbjct 129 LLRHYRAFGVSAILDYGVEEDLSPEEAEHK 158
> Hs10864043
Length=536
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 30/91 (32%), Positives = 43/91 (47%), Gaps = 0/91 (0%)
Query 109 AAALRLYSMSDLLRSLFVFSLCRHHWLVDNWESLLNVSCRIFGSRAVFAFIRFSFFKVFC 168
A L +L R+L V LC LV + L S R+ GSR AF+R S + F
Sbjct 102 GGAFHLKGTGELTRALLVLRLCAWPPLVTHGLLLQAWSRRLLGSRLSGAFLRASVYGQFV 161
Query 169 GGEALEEVVQTLDKLKQRNLGAILDYAAEQQ 199
GE EEV + +L+ +L +L E++
Sbjct 162 AGETAEEVKGCVQQLRTLSLRPLLAVPTEEE 192
> HsM5174645
Length=516
Score = 33.9 bits (76), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 18/63 (28%), Positives = 28/63 (44%), Gaps = 0/63 (0%)
Query 146 SCRIFGSRAVFAFIRFSFFKVFCGGEALEEVVQTLDKLKQRNLGAILDYAAEQQLPARAA 205
S ++ G R ++ +F+ F GE E + L + + AILDY E+ L A
Sbjct 12 SRKLLGQRLFNKLMKMTFYGHFVAGEDQESIQPLLRHYRAFGVSAILDYGVEEDLSPEEA 71
Query 206 ARK 208
K
Sbjct 72 EHK 74
> 7300817
Length=1329
Score = 32.3 bits (72), Expect = 0.73, Method: Composition-based stats.
Identities = 17/69 (24%), Positives = 33/69 (47%), Gaps = 2/69 (2%)
Query 20 AGARAATAAAYHVATPAASIAASRWLCSRGFASGVGQQFIFAPGQ--QRQQTHPQEESHQ 77
+GA A T++ PAA + A++++C +G ++ A Q + +P +++
Sbjct 193 SGAEAITSSVSSSGLPAAGLQAAQFICQGYMPTGPQRRLWHAENAVWQFDRNYPYNQAYS 252
Query 78 PPSPAPRAP 86
PP P P
Sbjct 253 PPYGIPMMP 261
> Hs20162576
Length=384
Score = 28.5 bits (62), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 24/104 (23%), Positives = 42/104 (40%), Gaps = 10/104 (9%)
Query 56 QQFIFAPGQQRQQTHPQEESHQPPSPAPRAPPNQRQQQKQQLQQLVLTGDAAGAAALRLY 115
Q F+ AP R++ PQEE+ + P+ AP A + + + +A+ A
Sbjct 28 QWFLQAPRSPREERSPQEETPEGPTDAPAA----DEPPSELVPGPPCVANASANATADFE 83
Query 116 SMSDLLRSLFVFSLCRHHWLVDNWESLLNVSCRIFGSRAVFAFI 159
+ ++ + CRH L L + + G R VF +
Sbjct 84 QLPARIQDFLRYRHCRHFPL------LWDAPAKCAGGRGVFLLL 121
Lambda K H
0.324 0.134 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3718707824
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40