bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0877_orf1
Length=91
Score E
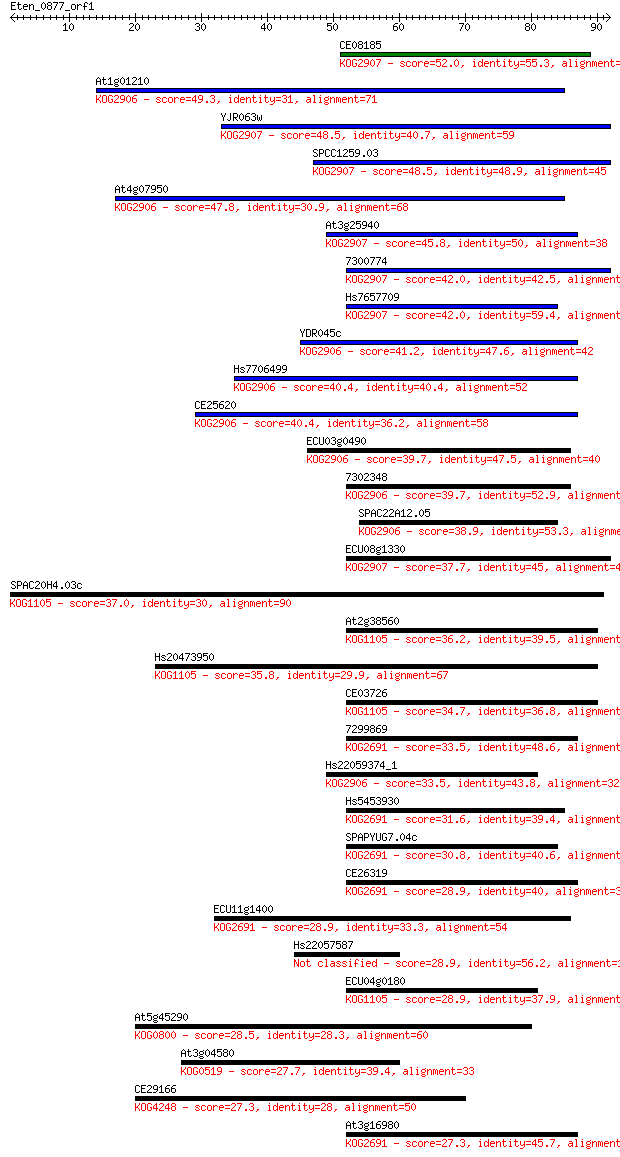
Sequences producing significant alignments: (Bits) Value
CE08185 52.0 3e-07
At1g01210 49.3 2e-06
YJR063w 48.5 3e-06
SPCC1259.03 48.5 3e-06
At4g07950 47.8 5e-06
At3g25940 45.8 2e-05
7300774 42.0 3e-04
Hs7657709 42.0 3e-04
YDR045c 41.2 5e-04
Hs7706499 40.4 7e-04
CE25620 40.4 8e-04
ECU03g0490 39.7 0.001
7302348 39.7 0.001
SPAC22A12.05 38.9 0.002
ECU08g1330 37.7 0.005
SPAC20H4.03c 37.0 0.009
At2g38560 36.2 0.015
Hs20473950 35.8 0.021
CE03726 34.7 0.045
7299869 33.5 0.10
Hs22059374_1 33.5 0.10
Hs5453930 31.6 0.35
SPAPYUG7.04c 30.8 0.70
CE26319 28.9 2.2
ECU11g1400 28.9 2.3
Hs22057587 28.9 2.4
ECU04g0180 28.9 2.4
At5g45290 28.5 3.1
At3g04580 27.7 6.1
CE29166 27.3 7.4
At3g16980 27.3 7.4
> CE08185
Length=119
Score = 52.0 bits (123), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 21/38 (55%), Positives = 27/38 (71%), Gaps = 0/38 (0%)
Query 51 ICERCGHNEAFFSTFQARSADEGMTVMYECTKCHHRRV 88
IC +CGH++A +ST Q RSADEG TV Y C KC + +
Sbjct 79 ICTKCGHSKASYSTMQTRSADEGQTVFYTCLKCKKKDI 116
> At1g01210
Length=106
Score = 49.3 bits (116), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 22/71 (30%), Positives = 39/71 (54%), Gaps = 0/71 (0%)
Query 14 SWQQRFLGAEFEQQLLQHHLKIMLSAGDGGNKTTCREICERCGHNEAFFSTFQARSADEG 73
++ QR + + +Q L++ ++ +++ D C RCGH++A+F + Q RSADE
Sbjct 30 AYIQRQVEIKKKQLLVKKSIEAVVTKDDIPTAAETEAPCPRCGHDKAYFKSMQIRSADEP 89
Query 74 MTVMYECTKCH 84
+ Y C KC
Sbjct 90 ESRFYRCLKCE 100
> YJR063w
Length=125
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 24/59 (40%), Positives = 33/59 (55%), Gaps = 0/59 (0%)
Query 33 LKIMLSAGDGGNKTTCREICERCGHNEAFFSTFQARSADEGMTVMYECTKCHHRRVFNN 91
+K L + + T +E C +CG+ E + T Q RSADEG TV Y CT C ++ NN
Sbjct 67 VKTSLKKNELKDGATIKEKCPQCGNEEMNYHTLQLRSADEGATVFYTCTSCGYKFRTNN 125
> SPCC1259.03
Length=119
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 22/45 (48%), Positives = 28/45 (62%), Gaps = 0/45 (0%)
Query 47 TCREICERCGHNEAFFSTFQARSADEGMTVMYECTKCHHRRVFNN 91
T E C +CG++ F T Q RSADEG TV YEC +C ++ NN
Sbjct 75 TIEEKCPKCGNDHMTFHTLQLRSADEGSTVFYECPRCAYKFSTNN 119
> At4g07950
Length=106
Score = 47.8 bits (112), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 21/68 (30%), Positives = 37/68 (54%), Gaps = 0/68 (0%)
Query 17 QRFLGAEFEQQLLQHHLKIMLSAGDGGNKTTCREICERCGHNEAFFSTFQARSADEGMTV 76
+R + + +Q L++ ++ +++ D C RCGH++A+F + Q RSADE +
Sbjct 33 ERRVEIKKKQLLVKKSIEPVVTKDDIPTAAETEAPCPRCGHDKAYFKSMQIRSADEPESR 92
Query 77 MYECTKCH 84
Y C KC
Sbjct 93 FYRCLKCE 100
> At3g25940
Length=91
Score = 45.8 bits (107), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 19/38 (50%), Positives = 24/38 (63%), Gaps = 0/38 (0%)
Query 49 REICERCGHNEAFFSTFQARSADEGMTVMYECTKCHHR 86
++ CE+C H E ++T Q RSADEG T Y C C HR
Sbjct 50 KKACEKCQHPELVYTTRQTRSADEGQTTYYTCPNCAHR 87
> 7300774
Length=120
Score = 42.0 bits (97), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 17/40 (42%), Positives = 25/40 (62%), Gaps = 0/40 (0%)
Query 52 CERCGHNEAFFSTFQARSADEGMTVMYECTKCHHRRVFNN 91
C +C H++ ++T Q RSADEG TV + C KC + N+
Sbjct 81 CPKCNHDKMSYATLQLRSADEGQTVFFTCLKCKFKESENS 120
> Hs7657709
Length=126
Score = 42.0 bits (97), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 19/32 (59%), Positives = 20/32 (62%), Gaps = 0/32 (0%)
Query 52 CERCGHNEAFFSTFQARSADEGMTVMYECTKC 83
C RCGH + T Q RSADEG TV Y CT C
Sbjct 87 CPRCGHEGMAYHTRQMRSADEGQTVFYTCTNC 118
> YDR045c
Length=110
Score = 41.2 bits (95), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 20/42 (47%), Positives = 23/42 (54%), Gaps = 1/42 (2%)
Query 45 KTTCREICERCGHNEAFFSTFQARSADEGMTVMYECTKCHHR 86
KT C + CG A+F Q RSADE MT Y+C C HR
Sbjct 66 KTQCPNY-DTCGGESAYFFQLQIRSADEPMTTFYKCVNCGHR 106
> Hs7706499
Length=108
Score = 40.4 bits (93), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 21/54 (38%), Positives = 28/54 (51%), Gaps = 2/54 (3%)
Query 35 IMLSAGDGGNKTTCREICERCGHNEAFFSTFQARSADEGMTVMYEC--TKCHHR 86
++ A N + E C +C H A+F Q RSADE MT Y+C +C HR
Sbjct 52 VLGGAAAWENVDSTAESCPKCEHPRAYFMQLQTRSADEPMTTFYKCCNAQCGHR 105
> CE25620
Length=108
Score = 40.4 bits (93), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 21/60 (35%), Positives = 26/60 (43%), Gaps = 2/60 (3%)
Query 29 LQHHLKIMLSAGDGGNKTTCREICERCGHNEAFFSTFQARSADEGMTVMYECTK--CHHR 86
L+ ++ G N E C C H A+F Q RSADE T+ Y C C HR
Sbjct 46 LKDIDDVLGGPGAWANAQVTDETCPVCSHGRAYFMQLQTRSADEPSTIFYRCADNACAHR 105
> ECU03g0490
Length=104
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 19/40 (47%), Positives = 23/40 (57%), Gaps = 1/40 (2%)
Query 46 TTCREICERCGHNEAFFSTFQARSADEGMTVMYECTKCHH 85
T C + CE CG E F Q RSADE MT+ Y+C +C
Sbjct 62 TKCGKRCE-CGSEEVSFVELQTRSADEPMTIFYKCIRCKK 100
> 7302348
Length=108
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 18/34 (52%), Positives = 21/34 (61%), Gaps = 2/34 (5%)
Query 52 CERCGHNEAFFSTFQARSADEGMTVMYECTKCHH 85
C CGH A+F Q RSADE MT Y+C C+H
Sbjct 69 CPTCGHKRAYFMQIQTRSADEPMTTFYKC--CNH 100
> SPAC22A12.05
Length=109
Score = 38.9 bits (89), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 16/30 (53%), Positives = 20/30 (66%), Gaps = 0/30 (0%)
Query 54 RCGHNEAFFSTFQARSADEGMTVMYECTKC 83
+C +N A+F Q RSADE M+ Y CTKC
Sbjct 73 KCDNNRAYFFQLQIRSADEPMSTFYRCTKC 102
> ECU08g1330
Length=99
Score = 37.7 bits (86), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 18/40 (45%), Positives = 23/40 (57%), Gaps = 1/40 (2%)
Query 52 CERCGHNEAFFSTFQARSADEGMTVMYECTKCHHRRVFNN 91
C CG E ++T Q RS DEG TV Y C KC +R+ +
Sbjct 61 CPACGAEEMMYNTAQLRSTDEGQTVFYSC-KCGYRQTVQS 99
> SPAC20H4.03c
Length=293
Score = 37.0 bits (84), Expect = 0.009, Method: Composition-based stats.
Identities = 27/90 (30%), Positives = 35/90 (38%), Gaps = 5/90 (5%)
Query 1 PLRPGNFGSVSKQSWQQRFLGAEFEQQLLQHHLKIMLSAGDGGNKTTCREICERCGHNEA 60
P R S S +R A+ EQ+ L H T C +C +
Sbjct 209 PQRLSTMTSAELASEDRRKEDAKLEQENLFH-----AQGAKPQKAVTDLFTCGKCKQKKV 263
Query 61 FFSTFQARSADEGMTVMYECTKCHHRRVFN 90
+ Q RSADE MT ECT C +R F+
Sbjct 264 SYYQMQTRSADEPMTTFCECTVCGNRWKFS 293
> At2g38560
Length=378
Score = 36.2 bits (82), Expect = 0.015, Method: Composition-based stats.
Identities = 15/38 (39%), Positives = 18/38 (47%), Gaps = 0/38 (0%)
Query 52 CERCGHNEAFFSTFQARSADEGMTVMYECTKCHHRRVF 89
C RCG + + Q RSADE MT C C + F
Sbjct 340 CGRCGQRKCTYYQMQTRSADEPMTTYVTCVNCDNHWKF 377
> Hs20473950
Length=348
Score = 35.8 bits (81), Expect = 0.021, Method: Composition-based stats.
Identities = 20/67 (29%), Positives = 29/67 (43%), Gaps = 1/67 (1%)
Query 23 EFEQQLLQHHLKIMLSAGDGGNKTTCREICERCGHNEAFFSTFQARSADEGMTVMYECTK 82
E + Q ++ A GG T + C +C ++ Q RSADE MT C +
Sbjct 282 ELRNAMTQEAIREHQMAKTGGTTTDLFQ-CSKCKKKNCTYNQVQTRSADEPMTTFVLCNE 340
Query 83 CHHRRVF 89
C +R F
Sbjct 341 CGNRWKF 347
> CE03726
Length=308
Score = 34.7 bits (78), Expect = 0.045, Method: Composition-based stats.
Identities = 14/38 (36%), Positives = 20/38 (52%), Gaps = 0/38 (0%)
Query 52 CERCGHNEAFFSTFQARSADEGMTVMYECTKCHHRRVF 89
C +CG ++ Q RS+DE MT C +C +R F
Sbjct 270 CGKCGKKNCTYTQLQTRSSDEPMTTFVFCLECGNRWKF 307
> 7299869
Length=129
Score = 33.5 bits (75), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 17/37 (45%), Positives = 20/37 (54%), Gaps = 2/37 (5%)
Query 52 CERCGHNEAFFSTFQARSADEGMTVMYECT--KCHHR 86
C +C H EA F Q R A+E M + Y CT C HR
Sbjct 90 CPKCSHREAVFFQAQTRRAEEEMRLYYVCTNQNCTHR 126
> Hs22059374_1
Length=42
Score = 33.5 bits (75), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 14/32 (43%), Positives = 18/32 (56%), Gaps = 0/32 (0%)
Query 49 REICERCGHNEAFFSTFQARSADEGMTVMYEC 80
+E C +C H A+F Q SADE M Y+C
Sbjct 2 KEPCLKCKHPRAYFMQLQTCSADELMITFYKC 33
> Hs5453930
Length=125
Score = 31.6 bits (70), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 13/33 (39%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 52 CERCGHNEAFFSTFQARSADEGMTVMYECTKCH 84
C++CGH EA F + A++ M + Y CT H
Sbjct 86 CQKCGHKEAVFFQSHSARAEDAMRLYYVCTAPH 118
> SPAPYUG7.04c
Length=113
Score = 30.8 bits (68), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 13/32 (40%), Positives = 17/32 (53%), Gaps = 0/32 (0%)
Query 52 CERCGHNEAFFSTFQARSADEGMTVMYECTKC 83
C RC +EA F +R D MT++Y C C
Sbjct 75 CPRCHQHEAVFYQTHSRRGDTMMTLIYVCVHC 106
> CE26319
Length=167
Score = 28.9 bits (63), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 14/37 (37%), Positives = 19/37 (51%), Gaps = 2/37 (5%)
Query 52 CERCGHNEAFFSTFQARSADEGMTVMYECT--KCHHR 86
C CG ++A F Q + A+E M + Y C C HR
Sbjct 128 CPVCGKSKAVFFQAQTKKAEEEMRLYYVCASQDCQHR 164
> ECU11g1400
Length=108
Score = 28.9 bits (63), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 18/59 (30%), Positives = 27/59 (45%), Gaps = 7/59 (11%)
Query 32 HLKIMLSAGDGGNKTTCREI---CERCGHNEAFFSTFQARSADE--GMTVMYECTKCHH 85
H I L A D T + C CG N+ + FQ R +E +++ Y C +C+H
Sbjct 49 HASIELYARDLVQDPTLPSVKINCGNCGFNKGLY--FQPRGGEEDVALSIFYLCCRCYH 105
> Hs22057587
Length=475
Score = 28.9 bits (63), Expect = 2.4, Method: Composition-based stats.
Identities = 9/16 (56%), Positives = 10/16 (62%), Gaps = 0/16 (0%)
Query 44 NKTTCREICERCGHNE 59
N+ CR IC CGH E
Sbjct 11 NRNVCRPICRECGHEE 26
> ECU04g0180
Length=257
Score = 28.9 bits (63), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 11/29 (37%), Positives = 14/29 (48%), Gaps = 0/29 (0%)
Query 52 CERCGHNEAFFSTFQARSADEGMTVMYEC 80
C +CG + + Q RS DE MT C
Sbjct 220 CSKCGERKCSYRQLQTRSGDEPMTTFVTC 248
> At5g45290
Length=546
Score = 28.5 bits (62), Expect = 3.1, Method: Composition-based stats.
Identities = 17/60 (28%), Positives = 27/60 (45%), Gaps = 1/60 (1%)
Query 20 LGAEFEQQLLQHHLKIMLSAGDGGNKTTCREICERCGHNEAFFSTFQARSADEGMTVMYE 79
LG+ FE + H +LS D + TCR + R G A AR++ + ++ E
Sbjct 377 LGSRFENVAVHHDRSCVLSGQDQAGRCTCRAVTNR-GSTTATTDETNARASISRIVLLAE 435
> At3g04580
Length=766
Score = 27.7 bits (60), Expect = 6.1, Method: Composition-based stats.
Identities = 13/33 (39%), Positives = 18/33 (54%), Gaps = 0/33 (0%)
Query 27 QLLQHHLKIMLSAGDGGNKTTCREICERCGHNE 59
QL+ + L +L DGG T R ICE G ++
Sbjct 486 QLVMYMLGYILDMTDGGKTVTFRVICEGTGTSQ 518
> CE29166
Length=1620
Score = 27.3 bits (59), Expect = 7.4, Method: Composition-based stats.
Identities = 14/50 (28%), Positives = 24/50 (48%), Gaps = 0/50 (0%)
Query 20 LGAEFEQQLLQHHLKIMLSAGDGGNKTTCREICERCGHNEAFFSTFQARS 69
+ +EFE Q+ H L G + + I E +NE FF+ F +++
Sbjct 1333 VASEFEAQIRAHIRDNYLVGRTGLSNSELHGIAENLANNEQFFAIFMSQN 1382
> At3g16980
Length=114
Score = 27.3 bits (59), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 16/37 (43%), Positives = 20/37 (54%), Gaps = 3/37 (8%)
Query 52 CERCGHNEAFFSTFQARSADEGMTVMYEC--TKCHHR 86
C +C H EA F AR +EGMT+ + C C HR
Sbjct 76 CSKCQHREAVFFQATAR-GEEGMTLFFVCCNPNCGHR 111
Lambda K H
0.323 0.134 0.434
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1174483934
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40