bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0882_orf2
Length=101
Score E
Sequences producing significant alignments: (Bits) Value
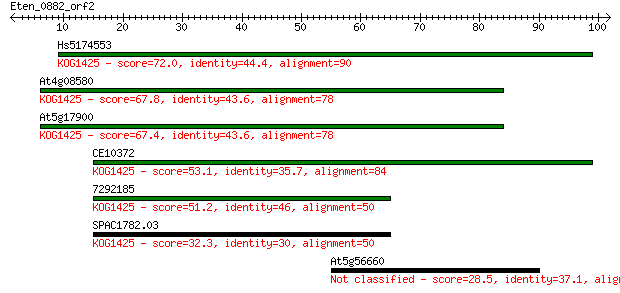
Hs5174553 72.0 3e-13
At4g08580 67.8 5e-12
At5g17900 67.4 6e-12
CE10372 53.1 1e-07
7292185 51.2 4e-07
SPAC1782.03 32.3 0.23
At5g56660 28.5 2.8
> Hs5174553
Length=439
Score = 72.0 bits (175), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 40/91 (43%), Positives = 54/91 (59%), Gaps = 7/91 (7%)
Query 9 EPLYLWDFNAPVGEDKVDKKTLPKILQLRRGELARGGRSKHTHLVDVDTSDLSHPWAQAT 68
E +Y DF+AP ED +K LPK++Q++ R GR+K+THLVD DT+ W Q +
Sbjct 353 EEVYKRDFSAPTLEDHFNKTILPKVMQVK--NFGRSGRTKYTHLVDQDTTSFDSAWGQES 410
Query 69 RDLQGQKLLK-KAAGIKGANDFERPSLRKPK 98
Q K K KAAG++ FERPS +K K
Sbjct 411 --AQNTKFFKQKAAGVRDV--FERPSAKKRK 437
> At4g08580
Length=435
Score = 67.8 bits (164), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 34/78 (43%), Positives = 52/78 (66%), Gaps = 4/78 (5%)
Query 6 SGEEPLYLWDFNAPVGEDKVDKKTLPKILQLRRGELARGGRSKHTHLVDVDTSDLSHPWA 65
+G + ++ DF+AP GED++DK LPK++Q++ R GR+K THLV+ DT+D S+PW
Sbjct 344 AGTDGIFQRDFSAPTGEDRLDKSILPKVMQVK--HFGRSGRTKWTHLVNEDTTDWSNPW- 400
Query 66 QATRDLQGQKLLKKAAGI 83
+ D +K KK AG+
Sbjct 401 -TSNDPLREKYNKKMAGM 417
> At5g17900
Length=435
Score = 67.4 bits (163), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 34/78 (43%), Positives = 52/78 (66%), Gaps = 4/78 (5%)
Query 6 SGEEPLYLWDFNAPVGEDKVDKKTLPKILQLRRGELARGGRSKHTHLVDVDTSDLSHPWA 65
+G + ++ DF+AP GED++DK LPK++Q++ R GR+K THLV+ DT+D S+PW
Sbjct 344 AGTDGIFQRDFSAPTGEDRLDKSILPKVMQVK--HFGRSGRTKWTHLVNEDTTDWSNPW- 400
Query 66 QATRDLQGQKLLKKAAGI 83
+ D +K KK AG+
Sbjct 401 -TSNDPLREKYNKKMAGM 417
> CE10372
Length=466
Score = 53.1 bits (126), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 30/84 (35%), Positives = 47/84 (55%), Gaps = 5/84 (5%)
Query 15 DFNAPVGEDKVDKKTLPKILQLRRGELARGGRSKHTHLVDVDTSDLSHPWAQATRDLQGQ 74
+F +D+ DK LPK++Q++ + R+K+THL + DT+D WA +T L Q
Sbjct 387 NFAEATNDDQFDKTILPKVMQVK--NFGKASRTKYTHLTEEDTTDHQGVWA-STNQLNSQ 443
Query 75 KLLKKAAGIKGANDFERPSLRKPK 98
K+A G + FERP+ +K K
Sbjct 444 FSTKRAGGSRPV--FERPATKKRK 465
> 7292185
Length=478
Score = 51.2 bits (121), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 23/50 (46%), Positives = 30/50 (60%), Gaps = 2/50 (4%)
Query 15 DFNAPVGEDKVDKKTLPKILQLRRGELARGGRSKHTHLVDVDTSDLSHPW 64
DF ED DK LPK++Q++ R GR+K+THLVD DT+ PW
Sbjct 397 DFAQATLEDHFDKTILPKVMQVK--NFGRCGRTKYTHLVDQDTTKFDSPW 444
> SPAC1782.03
Length=355
Score = 32.3 bits (72), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 15/50 (30%), Positives = 28/50 (56%), Gaps = 0/50 (0%)
Query 15 DFNAPVGEDKVDKKTLPKILQLRRGELARGGRSKHTHLVDVDTSDLSHPW 64
D++ + ++K LPK +Q+R A+ G+++ THL + DT+ W
Sbjct 276 DYSEATEGEVLNKDLLPKPMQIRGDLFAKAGQTRWTHLANEDTTKEGSAW 325
> At5g56660
Length=439
Score = 28.5 bits (62), Expect = 2.8, Method: Composition-based stats.
Identities = 13/35 (37%), Positives = 20/35 (57%), Gaps = 0/35 (0%)
Query 55 VDTSDLSHPWAQATRDLQGQKLLKKAAGIKGANDF 89
V+ DL + + RDL GQ+ +AA + G+ DF
Sbjct 336 VNNKDLYKQFKKVVRDLLGQEAFVEAAPVMGSEDF 370
Lambda K H
0.313 0.133 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1184494980
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40