bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
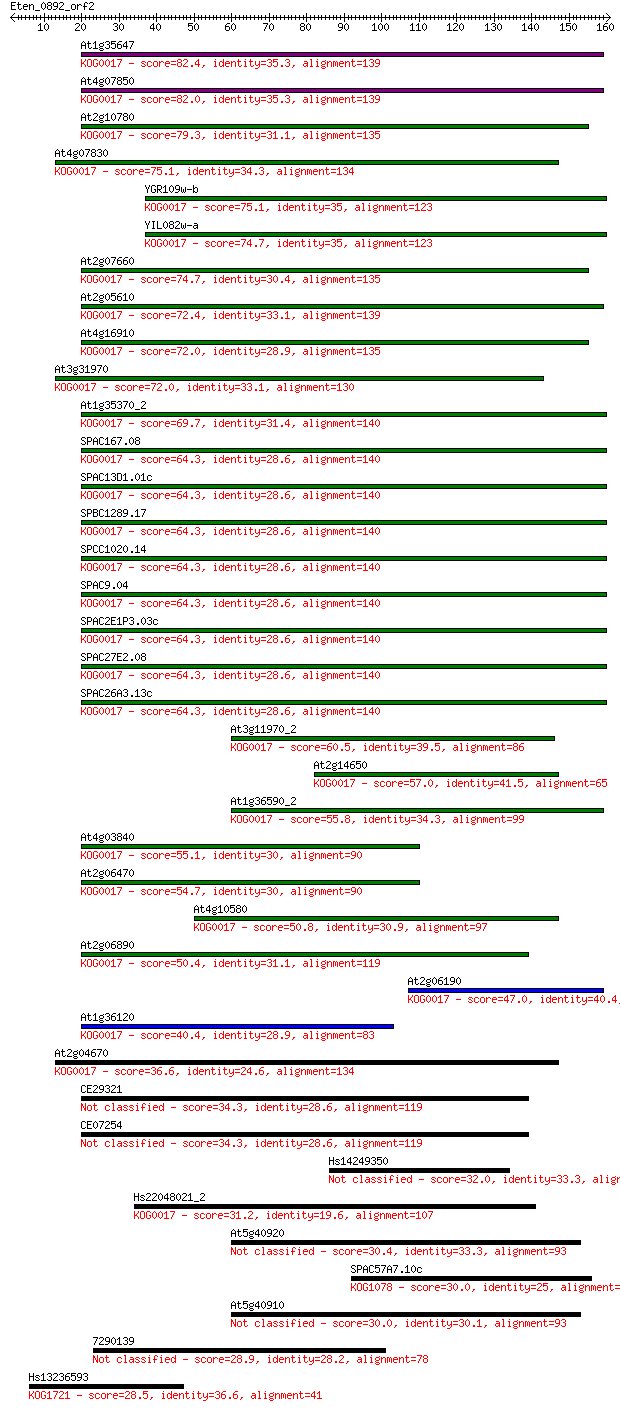
Query= Eten_0892_orf2
Length=160
Score E
Sequences producing significant alignments: (Bits) Value
At1g35647 82.4 4e-16
At4g07850 82.0 4e-16
At2g10780 79.3 3e-15
At4g07830 75.1 5e-14
YGR109w-b 75.1 6e-14
YIL082w-a 74.7 6e-14
At2g07660 74.7 7e-14
At2g05610 72.4 3e-13
At4g16910 72.0 5e-13
At3g31970 72.0 5e-13
At1g35370_2 69.7 2e-12
SPAC167.08 64.3 8e-11
SPAC13D1.01c 64.3 9e-11
SPBC1289.17 64.3 9e-11
SPCC1020.14 64.3 9e-11
SPAC9.04 64.3 9e-11
SPAC2E1P3.03c 64.3 9e-11
SPAC27E2.08 64.3 9e-11
SPAC26A3.13c 64.3 9e-11
At3g11970_2 60.5 2e-09
At2g14650 57.0 1e-08
At1g36590_2 55.8 3e-08
At4g03840 55.1 5e-08
At2g06470 54.7 8e-08
At4g10580 50.8 1e-06
At2g06890 50.4 1e-06
At2g06190 47.0 2e-05
At1g36120 40.4 0.001
At2g04670 36.6 0.022
CE29321 34.3 0.096
CE07254 34.3 0.096
Hs14249350 32.0 0.48
Hs22048021_2 31.2 0.95
At5g40920 30.4 1.4
SPAC57A7.10c 30.0 1.9
At5g40910 30.0 2.0
7290139 28.9 4.6
Hs13236593 28.5 5.2
> At1g35647
Length=1495
Score = 82.4 bits (202), Expect = 4e-16, Method: Composition-based stats.
Identities = 49/139 (35%), Positives = 65/139 (46%), Gaps = 2/139 (1%)
Query 20 RICVPQFPEFLTQTLHSHHDHVTAGHGGQKKTFATLSKHQYWPGMLAYTTADVESSTHSR 79
R+C+P + H GH G KT + H +WP M E +
Sbjct 983 RLCIPN-SSLRELFIREAHGGGLMGHFGVSKTIKVMQDHFHWPHMKRDVERICERCPTCK 1041
Query 80 ASKSLNQKPAGLLRQLLIPSRRWAHVSLDFITDLPLTMTGHDSILVMVDSLSKMAHFFPA 139
+K+ +Q P GL L IPS W +S+DF+ LP T TG DSI V+VD SKMAHF P
Sbjct 1042 QAKAKSQ-PHGLYTPLPIPSHPWNDISMDFVVGLPRTRTGKDSIFVVVDRFSKMAHFIPC 1100
Query 140 KKSFTAADTVELLADCLIR 158
K+ A L ++R
Sbjct 1101 HKTDDAIHIANLFFREVVR 1119
> At4g07850
Length=1138
Score = 82.0 bits (201), Expect = 4e-16, Method: Composition-based stats.
Identities = 49/139 (35%), Positives = 65/139 (46%), Gaps = 2/139 (1%)
Query 20 RICVPQFPEFLTQTLHSHHDHVTAGHGGQKKTFATLSKHQYWPGMLAYTTADVESSTHSR 79
R+C+P + H GH G KT + H +WP M E T +
Sbjct 750 RLCIPN-SSLRELFIREAHGGGLMGHFGVSKTLKVMQDHFHWPHMKRDVERMCERCTTCK 808
Query 80 ASKSLNQKPAGLLRQLLIPSRRWAHVSLDFITDLPLTMTGHDSILVMVDSLSKMAHFFPA 139
+K+ +Q P GL L IP W +S+DF+ LP T TG DSI V+VD SKMAHF P
Sbjct 809 QAKAKSQ-PHGLCTPLPIPLHPWNDISMDFVVGLPRTRTGKDSIFVVVDRFSKMAHFIPC 867
Query 140 KKSFTAADTVELLADCLIR 158
K+ A L ++R
Sbjct 868 HKTDDAMHIANLFFREVVR 886
> At2g10780
Length=1611
Score = 79.3 bits (194), Expect = 3e-15, Method: Composition-based stats.
Identities = 42/136 (30%), Positives = 71/136 (52%), Gaps = 5/136 (3%)
Query 20 RICVPQFPEFLTQTLHSHHDHVTAGHGGQKKTFATLSKHQYWPGMLAYTTADVESSTHSR 79
R+CVP + L H + H G K + L ++ +W GM V +
Sbjct 1150 RVCVPNDRALKEEILREAHQSKFSIHPGSNKMYRDLKRYYHWVGMKKDVARWVAKCPTCQ 1209
Query 80 ASKSLNQKPAGLLRQLLIPSRRWAHVSLDFITDLPLTM-TGHDSILVMVDSLSKMAHFFP 138
K+ +Q P+GLL+ L IP +W H+++DF+T LP + + H+++ V+VD L+K AHF
Sbjct 1210 LVKAEHQVPSGLLQNLPIPEWKWDHITMDFVTGLPTGIKSKHNAVWVVVDRLTKSAHFM- 1268
Query 139 AKKSFTAADTVELLAD 154
+ + D E++A+
Sbjct 1269 ---AISDKDGAEIIAE 1281
> At4g07830
Length=611
Score = 75.1 bits (183), Expect = 5e-14, Method: Composition-based stats.
Identities = 46/134 (34%), Positives = 69/134 (51%), Gaps = 3/134 (2%)
Query 13 IRVHGLWRICVPQFPEFLTQTLHSHHDHVTAGHGGQKKTFATLSKHQYWPGMLAYTTADV 72
I VHG R+CVP+ E + L H + + H G K + L ++ W GM V
Sbjct 155 ILVHG--RVCVPKDKELRQEILSEAHASMFSIHPGATKMYRDLKRYYQWVGMKRDVGNWV 212
Query 73 ESSTHSRASKSLNQKPAGLLRQLLIPSRRWAHVSLDFITDLPLTMTGHDSILVMVDSLSK 132
E + K +Q LL+ L IP +W +++DF+ LP++ T D+I V+VD L+K
Sbjct 213 EECDVCQLVKIEHQVSGSLLQSLPIPEWKWDFITMDFVVGLPVSRT-KDAIWVIVDRLTK 271
Query 133 MAHFFPAKKSFTAA 146
AHF +K+ AA
Sbjct 272 SAHFLAIRKTDGAA 285
> YGR109w-b
Length=1547
Score = 75.1 bits (183), Expect = 6e-14, Method: Composition-based stats.
Identities = 43/124 (34%), Positives = 61/124 (49%), Gaps = 1/124 (0%)
Query 37 HHDHVT-AGHGGQKKTFATLSKHQYWPGMLAYTTADVESSTHSRASKSLNQKPAGLLRQL 95
+HDH GH G T A +S YWP + + + + KS + GLL+ L
Sbjct 1103 YHDHTLFGGHFGVTVTLAKISPIYYWPKLQHSIIQYIRTCVQCQLIKSHRPRLHGLLQPL 1162
Query 96 LIPSRRWAHVSLDFITDLPLTMTGHDSILVMVDSLSKMAHFFPAKKSFTAADTVELLADC 155
I RW +S+DF+T LP T + ILV+VD SK AHF +K+ A ++LL
Sbjct 1163 PIAEGRWLDISMDFVTGLPPTSNNLNMILVVVDRFSKRAHFIATRKTLDATQLIDLLFRY 1222
Query 156 LIRY 159
+ Y
Sbjct 1223 IFSY 1226
> YIL082w-a
Length=1498
Score = 74.7 bits (182), Expect = 6e-14, Method: Composition-based stats.
Identities = 43/124 (34%), Positives = 61/124 (49%), Gaps = 1/124 (0%)
Query 37 HHDHVT-AGHGGQKKTFATLSKHQYWPGMLAYTTADVESSTHSRASKSLNQKPAGLLRQL 95
+HDH GH G T A +S YWP + + + + KS + GLL+ L
Sbjct 1129 YHDHTLFGGHFGVTVTLAKISPIYYWPKLQHSIIQYIRTCVQCQLIKSHRPRLHGLLQPL 1188
Query 96 LIPSRRWAHVSLDFITDLPLTMTGHDSILVMVDSLSKMAHFFPAKKSFTAADTVELLADC 155
I RW +S+DF+T LP T + ILV+VD SK AHF +K+ A ++LL
Sbjct 1189 PIAEGRWLDISMDFVTGLPPTSNNLNMILVVVDRFSKRAHFIATRKTLDATQLIDLLFRY 1248
Query 156 LIRY 159
+ Y
Sbjct 1249 IFSY 1252
> At2g07660
Length=949
Score = 74.7 bits (182), Expect = 7e-14, Method: Composition-based stats.
Identities = 41/136 (30%), Positives = 70/136 (51%), Gaps = 5/136 (3%)
Query 20 RICVPQFPEFLTQTLHSHHDHVTAGHGGQKKTFATLSKHQYWPGMLAYTTADVESSTHSR 79
R+CVP + L H + H G K + L ++ +W GM V +
Sbjct 542 RVCVPNDRALKEEILREAHQSKFSIHPGSNKMYRDLKRYYHWVGMRKDVARWVAKCPTCQ 601
Query 80 ASKSLNQKPAGLLRQLLIPSRRWAHVSLDFITDLPLTM-TGHDSILVMVDSLSKMAHFFP 138
K+ +Q P+GLL+ L I +W H+++DF+T LP + + H+++ V+VD L+K AHF
Sbjct 602 LVKAEHQVPSGLLQNLPISEWKWDHITMDFVTRLPTGIKSKHNAVWVVVDRLTKSAHFM- 660
Query 139 AKKSFTAADTVELLAD 154
+ + D E++A+
Sbjct 661 ---AISDKDGAEIIAE 673
> At2g05610
Length=780
Score = 72.4 bits (176), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 46/139 (33%), Positives = 64/139 (46%), Gaps = 1/139 (0%)
Query 20 RICVPQFPEFLTQTLHSHHDHVTAGHGGQKKTFATLSKHQYWPGMLAYTTADVESSTHSR 79
+I VP L H GH G++ T + YW M+ A + S +
Sbjct 462 KIVVPNNSGIKDTILRWLHCSGMGGHSGKEVTHQRVKGLFYWKSMVKDIQAFIRSCGTCQ 521
Query 80 ASKSLNQKPAGLLRQLLIPSRRWAHVSLDFITDLPLTMTGHDSILVMVDSLSKMAHFFPA 139
KS N GLL+ L IP R W+ VS+DFI LPL+ G I+V+VD LSK AHF
Sbjct 522 QCKSDNAASPGLLQPLPIPDRIWSDVSMDFIDGLPLS-NGKTVIMVVVDRLSKAAHFIAL 580
Query 140 KKSFTAADTVELLADCLIR 158
++A + D + +
Sbjct 581 AHPYSAMTVAQAYLDNVFK 599
> At4g16910
Length=687
Score = 72.0 bits (175), Expect = 5e-13, Method: Composition-based stats.
Identities = 39/136 (28%), Positives = 68/136 (50%), Gaps = 13/136 (9%)
Query 20 RICVPQFPEFLTQTLHSHHDHVTAGHGGQKKTFATLSKHQYWPGMLAYTTADVESSTHSR 79
R+C P + L H + H G K + L ++ +W GM +
Sbjct 242 RVCGPNDKALKEEILKEAHQSKFSIHPGSNKMYRDLKRYYHWVGM--------KKDVARW 293
Query 80 ASKSLNQKPAGLLRQLLIPSRRWAHVSLDFITDLPLTM-TGHDSILVMVDSLSKMAHFFP 138
+K +Q P+G+L+ L IP +W H+ +DF+T LP + + H+++ V+VD L+K AHF
Sbjct 294 VAKEEHQVPSGMLQNLPIPEWKWDHIMMDFVTGLPTGIKSKHNAVWVVVDRLTKSAHFM- 352
Query 139 AKKSFTAADTVELLAD 154
+ + D E++A+
Sbjct 353 ---AISDKDAAEIIAE 365
> At3g31970
Length=1329
Score = 72.0 bits (175), Expect = 5e-13, Method: Composition-based stats.
Identities = 43/130 (33%), Positives = 67/130 (51%), Gaps = 3/130 (2%)
Query 13 IRVHGLWRICVPQFPEFLTQTLHSHHDHVTAGHGGQKKTFATLSKHQYWPGMLAYTTADV 72
I VHG R+CVP+ E + L H + + H K + L ++ W GM V
Sbjct 888 ILVHG--RVCVPKDEELRREILSEAHASMFSIHPRATKMYRDLKRYYQWVGMKRDVANWV 945
Query 73 ESSTHSRASKSLNQKPAGLLRQLLIPSRRWAHVSLDFITDLPLTMTGHDSILVMVDSLSK 132
+ K+ +Q P GLL+ L I +W +++DF+ LP++ T D+I V+VD L+K
Sbjct 946 TECDVCQLVKAEHQVPGGLLQSLPILEWKWDFITMDFVVGLPVSRT-KDAIWVIVDRLTK 1004
Query 133 MAHFFPAKKS 142
AHF +K+
Sbjct 1005 SAHFLAIRKT 1014
> At1g35370_2
Length=923
Score = 69.7 bits (169), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 44/140 (31%), Positives = 63/140 (45%), Gaps = 1/140 (0%)
Query 20 RICVPQFPEFLTQTLHSHHDHVTAGHGGQKKTFATLSKHQYWPGMLAYTTADVESSTHSR 79
+I VP E + L H G G+ + + YW GM+ A + S +
Sbjct 518 KIVVPNDVEITNKLLQWLHCSGMGGRSGRDASHQRVKSLFYWKGMVKDIQAFIRSCGTCQ 577
Query 80 ASKSLNQKPAGLLRQLLIPSRRWAHVSLDFITDLPLTMTGHDSILVMVDSLSKMAHFFPA 139
KS N GLL+ L IP + W VS+DFI LP G I+V+VD LSK AHF
Sbjct 578 QCKSDNAAYPGLLQPLPIPDKIWCDVSMDFIEGLP-NSGGKSVIMVVVDRLSKAAHFVAL 636
Query 140 KKSFTAADTVELLADCLIRY 159
++A + D + ++
Sbjct 637 AHPYSALTVAQAFLDNVYKH 656
> SPAC167.08
Length=1214
Score = 64.3 bits (155), Expect = 8e-11, Method: Composition-based stats.
Identities = 40/140 (28%), Positives = 67/140 (47%), Gaps = 1/140 (0%)
Query 20 RICVPQFPEFLTQTLHSHHDHVTAGHGGQKKTFATLSKHQYWPGMLAYTTADVESSTHSR 79
+I +P + + +H+ H G + + + W G+ V++ +
Sbjct 783 QILLPNDTQLTRTIIKKYHEEGKLIHPGIELLTNIILRRFTWKGIRKQIQEYVQNCHTCQ 842
Query 80 ASKSLNQKPAGLLRQLLIPSRRWAHVSLDFITDLPLTMTGHDSILVMVDSLSKMAHFFPA 139
+KS N KP G L+ + R W +S+DFIT LP + +G++++ V+VD SKMA P
Sbjct 843 INKSRNHKPYGPLQPIPPSERPWESLSMDFITALPES-SGYNALFVVVDRFSKMAILVPC 901
Query 140 KKSFTAADTVELLADCLIRY 159
KS TA T + +I Y
Sbjct 902 TKSITAEQTARMFDQRVIAY 921
> SPAC13D1.01c
Length=1333
Score = 64.3 bits (155), Expect = 9e-11, Method: Composition-based stats.
Identities = 40/140 (28%), Positives = 67/140 (47%), Gaps = 1/140 (0%)
Query 20 RICVPQFPEFLTQTLHSHHDHVTAGHGGQKKTFATLSKHQYWPGMLAYTTADVESSTHSR 79
+I +P + + +H+ H G + + + W G+ V++ +
Sbjct 902 QILLPNDTQLTRTIIKKYHEEGKLIHPGIELLTNIILRRFTWKGIRKQIQEYVQNCHTCQ 961
Query 80 ASKSLNQKPAGLLRQLLIPSRRWAHVSLDFITDLPLTMTGHDSILVMVDSLSKMAHFFPA 139
+KS N KP G L+ + R W +S+DFIT LP + +G++++ V+VD SKMA P
Sbjct 962 INKSRNHKPYGPLQPIPPSERPWESLSMDFITALPES-SGYNALFVVVDRFSKMAILVPC 1020
Query 140 KKSFTAADTVELLADCLIRY 159
KS TA T + +I Y
Sbjct 1021 TKSITAEQTARMFDQRVIAY 1040
> SPBC1289.17
Length=1333
Score = 64.3 bits (155), Expect = 9e-11, Method: Composition-based stats.
Identities = 40/140 (28%), Positives = 67/140 (47%), Gaps = 1/140 (0%)
Query 20 RICVPQFPEFLTQTLHSHHDHVTAGHGGQKKTFATLSKHQYWPGMLAYTTADVESSTHSR 79
+I +P + + +H+ H G + + + W G+ V++ +
Sbjct 902 QILLPNDTQLTRTIIKKYHEEGKLIHPGIELLTNIILRRFTWKGIRKQIQEYVQNCHTCQ 961
Query 80 ASKSLNQKPAGLLRQLLIPSRRWAHVSLDFITDLPLTMTGHDSILVMVDSLSKMAHFFPA 139
+KS N KP G L+ + R W +S+DFIT LP + +G++++ V+VD SKMA P
Sbjct 962 INKSRNHKPYGPLQPIPPSERPWESLSMDFITALPES-SGYNALFVVVDRFSKMAILVPC 1020
Query 140 KKSFTAADTVELLADCLIRY 159
KS TA T + +I Y
Sbjct 1021 TKSITAEQTARMFDQRVIAY 1040
> SPCC1020.14
Length=1333
Score = 64.3 bits (155), Expect = 9e-11, Method: Composition-based stats.
Identities = 40/140 (28%), Positives = 67/140 (47%), Gaps = 1/140 (0%)
Query 20 RICVPQFPEFLTQTLHSHHDHVTAGHGGQKKTFATLSKHQYWPGMLAYTTADVESSTHSR 79
+I +P + + +H+ H G + + + W G+ V++ +
Sbjct 902 QILLPNDTQLTRTIIKKYHEEGKLIHPGIELLTNIILRRFTWKGIRKQIQEYVQNCHTCQ 961
Query 80 ASKSLNQKPAGLLRQLLIPSRRWAHVSLDFITDLPLTMTGHDSILVMVDSLSKMAHFFPA 139
+KS N KP G L+ + R W +S+DFIT LP + +G++++ V+VD SKMA P
Sbjct 962 INKSRNHKPYGPLQPIPPSERPWESLSMDFITALPES-SGYNALFVVVDRFSKMAILVPC 1020
Query 140 KKSFTAADTVELLADCLIRY 159
KS TA T + +I Y
Sbjct 1021 TKSITAEQTARMFDQRVIAY 1040
> SPAC9.04
Length=1333
Score = 64.3 bits (155), Expect = 9e-11, Method: Composition-based stats.
Identities = 40/140 (28%), Positives = 67/140 (47%), Gaps = 1/140 (0%)
Query 20 RICVPQFPEFLTQTLHSHHDHVTAGHGGQKKTFATLSKHQYWPGMLAYTTADVESSTHSR 79
+I +P + + +H+ H G + + + W G+ V++ +
Sbjct 902 QILLPNDTQLTRTIIKKYHEEGKLIHPGIELLTNIILRRFTWKGIRKQIQEYVQNCHTCQ 961
Query 80 ASKSLNQKPAGLLRQLLIPSRRWAHVSLDFITDLPLTMTGHDSILVMVDSLSKMAHFFPA 139
+KS N KP G L+ + R W +S+DFIT LP + +G++++ V+VD SKMA P
Sbjct 962 INKSRNHKPYGPLQPIPPSERPWESLSMDFITALPES-SGYNALFVVVDRFSKMAILVPC 1020
Query 140 KKSFTAADTVELLADCLIRY 159
KS TA T + +I Y
Sbjct 1021 TKSITAEQTARMFDQRVIAY 1040
> SPAC2E1P3.03c
Length=1333
Score = 64.3 bits (155), Expect = 9e-11, Method: Composition-based stats.
Identities = 40/140 (28%), Positives = 67/140 (47%), Gaps = 1/140 (0%)
Query 20 RICVPQFPEFLTQTLHSHHDHVTAGHGGQKKTFATLSKHQYWPGMLAYTTADVESSTHSR 79
+I +P + + +H+ H G + + + W G+ V++ +
Sbjct 902 QILLPNDTQLTRTIIKKYHEEGKLIHPGIELLTNIILRRFTWKGIRKQIQEYVQNCHTCQ 961
Query 80 ASKSLNQKPAGLLRQLLIPSRRWAHVSLDFITDLPLTMTGHDSILVMVDSLSKMAHFFPA 139
+KS N KP G L+ + R W +S+DFIT LP + +G++++ V+VD SKMA P
Sbjct 962 INKSRNHKPYGPLQPIPPSERPWESLSMDFITALPES-SGYNALFVVVDRFSKMAILVPC 1020
Query 140 KKSFTAADTVELLADCLIRY 159
KS TA T + +I Y
Sbjct 1021 TKSITAEQTARMFDQRVIAY 1040
> SPAC27E2.08
Length=1333
Score = 64.3 bits (155), Expect = 9e-11, Method: Composition-based stats.
Identities = 40/140 (28%), Positives = 67/140 (47%), Gaps = 1/140 (0%)
Query 20 RICVPQFPEFLTQTLHSHHDHVTAGHGGQKKTFATLSKHQYWPGMLAYTTADVESSTHSR 79
+I +P + + +H+ H G + + + W G+ V++ +
Sbjct 902 QILLPNDTQLTRTIIKKYHEEGKLIHPGIELLTNIILRRFTWKGIRKQIQEYVQNCHTCQ 961
Query 80 ASKSLNQKPAGLLRQLLIPSRRWAHVSLDFITDLPLTMTGHDSILVMVDSLSKMAHFFPA 139
+KS N KP G L+ + R W +S+DFIT LP + +G++++ V+VD SKMA P
Sbjct 962 INKSRNHKPYGPLQPIPPSERPWESLSMDFITALPES-SGYNALFVVVDRFSKMAILVPC 1020
Query 140 KKSFTAADTVELLADCLIRY 159
KS TA T + +I Y
Sbjct 1021 TKSITAEQTARMFDQRVIAY 1040
> SPAC26A3.13c
Length=1333
Score = 64.3 bits (155), Expect = 9e-11, Method: Composition-based stats.
Identities = 40/140 (28%), Positives = 67/140 (47%), Gaps = 1/140 (0%)
Query 20 RICVPQFPEFLTQTLHSHHDHVTAGHGGQKKTFATLSKHQYWPGMLAYTTADVESSTHSR 79
+I +P + + +H+ H G + + + W G+ V++ +
Sbjct 902 QILLPNDTQLTRTIIKKYHEEGKLIHPGIELLTNIILRRFTWKGIRKQIQEYVQNCHTCQ 961
Query 80 ASKSLNQKPAGLLRQLLIPSRRWAHVSLDFITDLPLTMTGHDSILVMVDSLSKMAHFFPA 139
+KS N KP G L+ + R W +S+DFIT LP + +G++++ V+VD SKMA P
Sbjct 962 INKSRNHKPYGPLQPIPPSERPWESLSMDFITALPES-SGYNALFVVVDRFSKMAILVPC 1020
Query 140 KKSFTAADTVELLADCLIRY 159
KS TA T + +I Y
Sbjct 1021 TKSITAEQTARMFDQRVIAY 1040
> At3g11970_2
Length=958
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 34/86 (39%), Positives = 46/86 (53%), Gaps = 1/86 (1%)
Query 60 YWPGMLAYTTADVESSTHSRASKSLNQKPAGLLRQLLIPSRRWAHVSLDFITDLPLTMTG 119
YW GM+ A + S + KS GLL+ L IP W+ VS+DFI LP++ G
Sbjct 570 YWKGMIKDIQAYIRSCGTCQQCKSDPAASPGLLQPLPIPDTIWSEVSMDFIEGLPVS-GG 628
Query 120 HDSILVMVDSLSKMAHFFPAKKSFTA 145
I+V+VD LSK AHF ++A
Sbjct 629 KTVIMVVVDRLSKAAHFIALSHPYSA 654
> At2g14650
Length=1328
Score = 57.0 bits (136), Expect = 1e-08, Method: Composition-based stats.
Identities = 27/65 (41%), Positives = 43/65 (66%), Gaps = 1/65 (1%)
Query 82 KSLNQKPAGLLRQLLIPSRRWAHVSLDFITDLPLTMTGHDSILVMVDSLSKMAHFFPAKK 141
K+ +Q P G+L+ L IP +W +++DF+ LP++ T D+I V+VD L+K AHF +K
Sbjct 957 KAEHQVPGGMLQSLPIPEWKWDFITIDFVVGLPVSRT-KDAIWVIVDRLTKSAHFLAIRK 1015
Query 142 SFTAA 146
+ AA
Sbjct 1016 TDGAA 1020
> At1g36590_2
Length=958
Score = 55.8 bits (133), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 34/99 (34%), Positives = 49/99 (49%), Gaps = 1/99 (1%)
Query 60 YWPGMLAYTTADVESSTHSRASKSLNQKPAGLLRQLLIPSRRWAHVSLDFITDLPLTMTG 119
Y GM+ A + S + KS GLL+ L IP W+ VS+DFI LP++ G
Sbjct 570 YSKGMIKDIQAYIRSCGTCQQCKSDPAASPGLLQPLPIPDTIWSEVSMDFIEGLPVS-GG 628
Query 120 HDSILVMVDSLSKMAHFFPAKKSFTAADTVELLADCLIR 158
I+V+VD LSK AHF ++A + D + +
Sbjct 629 KTVIMVVVDRLSKAAHFIALSHPYSALTVAQAYLDNVFK 667
> At4g03840
Length=973
Score = 55.1 bits (131), Expect = 5e-08, Method: Composition-based stats.
Identities = 27/90 (30%), Positives = 43/90 (47%), Gaps = 0/90 (0%)
Query 20 RICVPQFPEFLTQTLHSHHDHVTAGHGGQKKTFATLSKHQYWPGMLAYTTADVESSTHSR 79
R+CVP + L H + H G K + L ++ +W GM V +
Sbjct 600 RVCVPNNRALKEEILREAHQSKFSIHPGSNKIYRDLKRYYHWVGMKKDVARWVAKCPTCQ 659
Query 80 ASKSLNQKPAGLLRQLLIPSRRWAHVSLDF 109
K+ +Q P+GLL+ L IP +W H+++DF
Sbjct 660 LVKAEHQVPSGLLQNLPIPEWKWDHITMDF 689
> At2g06470
Length=899
Score = 54.7 bits (130), Expect = 8e-08, Method: Composition-based stats.
Identities = 27/90 (30%), Positives = 43/90 (47%), Gaps = 0/90 (0%)
Query 20 RICVPQFPEFLTQTLHSHHDHVTAGHGGQKKTFATLSKHQYWPGMLAYTTADVESSTHSR 79
R+CVP + L H + H G K + L ++ +W GM V +
Sbjct 600 RVCVPNDRALKEEILREAHQSKFSIHPGSNKMYRDLKRYYHWVGMKKDVARWVAKCPTCQ 659
Query 80 ASKSLNQKPAGLLRQLLIPSRRWAHVSLDF 109
K+ +Q P+GLL+ L IP +W H+++DF
Sbjct 660 LVKAEHQVPSGLLQNLPIPEWKWDHITMDF 689
> At4g10580
Length=1240
Score = 50.8 bits (120), Expect = 1e-06, Method: Composition-based stats.
Identities = 30/97 (30%), Positives = 50/97 (51%), Gaps = 1/97 (1%)
Query 50 KTFATLSKHQYWPGMLAYTTADVESSTHSRASKSLNQKPAGLLRQLLIPSRRWAHVSLDF 109
K + L ++ W GM V + K+ +Q G+L+ L IP +W +++D
Sbjct 960 KMYRDLKRYYQWVGMKMDVANWVAECDVCQLVKAEHQVLGGMLQSLPIPEWKWDFITMDL 1019
Query 110 ITDLPLTMTGHDSILVMVDSLSKMAHFFPAKKSFTAA 146
+ L ++ T D+I V+VD L+K AHF +K+ AA
Sbjct 1020 VVGLRVSRT-KDAIWVIVDRLTKSAHFLAIRKTDGAA 1055
> At2g06890
Length=1215
Score = 50.4 bits (119), Expect = 1e-06, Method: Composition-based stats.
Identities = 37/119 (31%), Positives = 49/119 (41%), Gaps = 4/119 (3%)
Query 20 RICVPQFPEFLTQTLHSHHDHVTAGHGGQKKTFATLSKHQYWPGMLAYTTADVESSTHSR 79
R+CVP + H GH G KT +++H WP M +
Sbjct 884 RLCVPNC-SLRDLFVREAHGGGLMGHFGIAKTLEVMTEHFRWPHMKCDVKRICGRCNTCK 942
Query 80 ASKSLNQKPAGLLRQLLIPSRRWAHVSLDFITDLPLTMTGHDSILVMVDSLSKMAHFFP 138
+KS Q P GL L IP W +S+DF+ LP TG DSI V+ + F P
Sbjct 943 QAKSKIQ-PNGLYTPLPIPKHPWNDISMDFVMGLP--RTGKDSIFVVYSPFQIVYGFNP 998
> At2g06190
Length=280
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 21/52 (40%), Positives = 31/52 (59%), Gaps = 0/52 (0%)
Query 107 LDFITDLPLTMTGHDSILVMVDSLSKMAHFFPAKKSFTAADTVELLADCLIR 158
+DF+ LP T G DS+ V+VD SKM HF KK+ A++ +L ++R
Sbjct 1 MDFVLGLPRTQRGVDSVFVVVDRFSKMTHFIACKKTADASNIAKLFFKEVVR 52
> At1g36120
Length=1235
Score = 40.4 bits (93), Expect = 0.001, Method: Composition-based stats.
Identities = 24/83 (28%), Positives = 36/83 (43%), Gaps = 2/83 (2%)
Query 20 RICVPQFPEFLTQTLHSHHDHVTAGHGGQKKTFATLSKHQYWPGMLAYTTADVESSTHSR 79
R+C+P+ E + L H + + H G K + L +H W GM V +
Sbjct 866 RVCLPKDEELRREILSEAHASMFSIHPGATKMYRDLKRHYQWVGMKRDVANWVTECDVCQ 925
Query 80 ASKSLNQKPAGLLRQLLIPSRRW 102
K+ +Q P GLL+ L P W
Sbjct 926 LVKAEHQVPGGLLQSL--PISEW 946
> At2g04670
Length=1411
Score = 36.6 bits (83), Expect = 0.022, Method: Composition-based stats.
Identities = 33/134 (24%), Positives = 51/134 (38%), Gaps = 34/134 (25%)
Query 13 IRVHGLWRICVPQFPEFLTQTLHSHHDHVTAGHGGQKKTFATLSKHQYWPGMLAYTTADV 72
I V+G R+CVP+ E + L H + + H G K + L ++ W GM V
Sbjct 1001 IFVYG--RVCVPKDEELRREILSEAHASMFSIHPGATKMYRDLKRYYQWVGMKRDVANWV 1058
Query 73 ESSTHSRASKSLNQKPAGLLRQLLIPSRRWAHVSLDFITDLPLTMTGHDSILVMVDSLSK 132
+ K+ +Q P D+I V++D L+K
Sbjct 1059 AECDVCQLVKAEHQVP--------------------------------DAIWVIMDRLTK 1086
Query 133 MAHFFPAKKSFTAA 146
AHF +K+ AA
Sbjct 1087 SAHFLAIRKTDGAA 1100
> CE29321
Length=2186
Score = 34.3 bits (77), Expect = 0.096, Method: Composition-based stats.
Identities = 34/126 (26%), Positives = 55/126 (43%), Gaps = 19/126 (15%)
Query 20 RICVPQFPEFLTQTLHSHHDHVTAGHGGQKKTFATLSKHQYWPGMLAYTTADVES----- 74
R VP+ + T L H+ + AGH G KK + + + YWP M V +
Sbjct 1457 RSVVPE--KIRTPLLKELHEGMLAGHFGIKKMWRMVHRKFYWPQMRVCVENCVRTCAKCL 1514
Query 75 --STHSRASKSLNQKPAGLLRQLLIPSRRWAHVSLDFITDLPLTMTGHDSILVMVDSLSK 132
+ HS+ + SL P + L I V+ D + D+ L++ G+ IL ++D +K
Sbjct 1515 CANDHSKLTSSLT--PYRMTFPLEI-------VACDLM-DVGLSVQGNRYILTIIDLFTK 1564
Query 133 MAHFFP 138
P
Sbjct 1565 YGTAVP 1570
> CE07254
Length=2175
Score = 34.3 bits (77), Expect = 0.096, Method: Composition-based stats.
Identities = 34/126 (26%), Positives = 55/126 (43%), Gaps = 19/126 (15%)
Query 20 RICVPQFPEFLTQTLHSHHDHVTAGHGGQKKTFATLSKHQYWPGMLAYTTADVES----- 74
R VP+ + T L H+ + AGH G KK + + + YWP M V +
Sbjct 1446 RSVVPE--KIRTPLLKELHEGMLAGHFGIKKMWRMVHRKFYWPQMRVCVENCVRTCAKCL 1503
Query 75 --STHSRASKSLNQKPAGLLRQLLIPSRRWAHVSLDFITDLPLTMTGHDSILVMVDSLSK 132
+ HS+ + SL P + L I V+ D + D+ L++ G+ IL ++D +K
Sbjct 1504 CANDHSKLTSSLT--PYRMTFPLEI-------VACDLM-DVGLSVQGNRYILTIIDLFTK 1553
Query 133 MAHFFP 138
P
Sbjct 1554 YGTAVP 1559
> Hs14249350
Length=538
Score = 32.0 bits (71), Expect = 0.48, Method: Composition-based stats.
Identities = 16/48 (33%), Positives = 28/48 (58%), Gaps = 1/48 (2%)
Query 86 QKPAGLLRQLLIPSRRWAHVSLDFITDLPLTMTGHDSILVMVDSLSKM 133
+ P G Q L+P W +V++D + + PL + GH ++ V V+S K+
Sbjct 316 RAPGGDPCQPLVPPLSWENVTVDKVLEFPL-LKGHPNLCVQVNSSEKL 362
> Hs22048021_2
Length=835
Score = 31.2 bits (69), Expect = 0.95, Method: Compositional matrix adjust.
Identities = 21/112 (18%), Positives = 46/112 (41%), Gaps = 9/112 (8%)
Query 34 LHSHHDHVTAGHGGQKKTFATLSKHQYWPGMLAYTTADVESSTHSRASKSLNQKPAGLLR 93
+ S HD H ++T+ L +WPGM + S + + L+
Sbjct 482 IFSVHDIPLGAHQRPEETYKKLRLLGWWPGMQEHVKDYCRSCLFCIPRNLIGSE----LK 537
Query 94 QLLIP-----SRRWAHVSLDFITDLPLTMTGHDSILVMVDSLSKMAHFFPAK 140
+ P + W+++ ++ + + ++ GH +L++ D ++ FP K
Sbjct 538 VIESPWPLRSTAPWSNLQIEVVGPVTISEEGHKHVLIVADPNTRWVEAFPLK 589
> At5g40920
Length=1066
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 31/109 (28%), Positives = 49/109 (44%), Gaps = 16/109 (14%)
Query 60 YWPGMLAYTTADVESSTHSRASKSL-NQKPAGLL----------RQLLI-----PSRRWA 103
Y MLA +T DVE+ + A+KSL + GL+ RQ+++ P +R
Sbjct 425 YVSTMLADSTLDVENGLKTLAAKSLVHISTHGLVRMHCLLQQLGRQVVVQQSGEPGKRQF 484
Query 104 HVSLDFITDLPLTMTGHDSILVMVDSLSKMAHFFPAKKSFTAADTVELL 152
V I D+ TG SI+ + +SK+ F K+ F ++ L
Sbjct 485 LVEAKEIRDVLANETGTGSIIGISFDMSKIGEFSIRKRVFEGMHNLKFL 533
> SPAC57A7.10c
Length=905
Score = 30.0 bits (66), Expect = 1.9, Method: Composition-based stats.
Identities = 16/64 (25%), Positives = 29/64 (45%), Gaps = 0/64 (0%)
Query 92 LRQLLIPSRRWAHVSLDFITDLPLTMTGHDSILVMVDSLSKMAHFFPAKKSFTAADTVEL 151
+R L + R L F++++ G++ VD++S M + P K A+ E
Sbjct 403 IRSLCLKFPRKQDSMLTFLSNILCDEGGYEFKRAAVDAISDMIKYIPESKERALAELCEF 462
Query 152 LADC 155
+ DC
Sbjct 463 IEDC 466
> At5g40910
Length=1104
Score = 30.0 bits (66), Expect = 2.0, Method: Composition-based stats.
Identities = 28/106 (26%), Positives = 48/106 (45%), Gaps = 13/106 (12%)
Query 60 YWPGMLAYTTADVESSTHSRASKSLNQKPA---------GLLRQLLI----PSRRWAHVS 106
Y MLA +T DVE+ + A+KSL L RQ+++ P +R V
Sbjct 442 YVTTMLADSTLDVENGLKTLAAKSLVSTNGWITMHCLLQQLGRQVVVQQGDPGKRQFLVE 501
Query 107 LDFITDLPLTMTGHDSILVMVDSLSKMAHFFPAKKSFTAADTVELL 152
I D+ TG +S++ + +SK+ +K++F ++ L
Sbjct 502 AKEIRDVLANETGTESVIGISFDISKIETLSISKRAFNRMRNLKFL 547
> 7290139
Length=2118
Score = 28.9 bits (63), Expect = 4.6, Method: Composition-based stats.
Identities = 22/79 (27%), Positives = 32/79 (40%), Gaps = 1/79 (1%)
Query 23 VPQFPEFLTQTLHSHHDHVTAG-HGGQKKTFATLSKHQYWPGMLAYTTADVESSTHSRAS 81
V Q P+ TQTL ++ T + TLS Q WPG++ ++ +V S
Sbjct 758 VVQAPKEFTQTLSANAVDTTGTINAVYPHDVITLSDSQEWPGLVTFSHLEVSEELELNGS 817
Query 82 KSLNQKPAGLLRQLLIPSR 100
Q L L+ SR
Sbjct 818 AQGRQFEEAPLNPTLLESR 836
> Hs13236593
Length=375
Score = 28.5 bits (62), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 15/41 (36%), Positives = 20/41 (48%), Gaps = 0/41 (0%)
Query 6 YLQPYLHIRVHGLWRICVPQFPEFLTQTLHSHHDHVTAGHG 46
YL+P+L I C F EF TQ++ + H V G G
Sbjct 250 YLRPHLRIHTGEKPYKCNQCFREFRTQSIFTRHKRVHTGEG 290
Lambda K H
0.325 0.135 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2172509160
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40