bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0897_orf1
Length=202
Score E
Sequences producing significant alignments: (Bits) Value
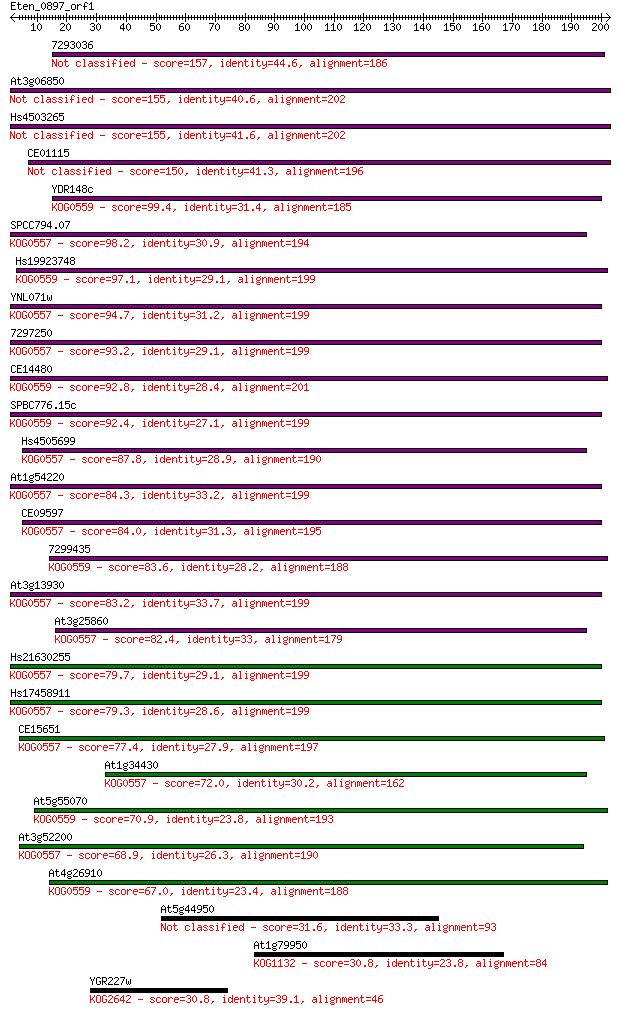
7293036 157 9e-39
At3g06850 155 4e-38
Hs4503265 155 5e-38
CE01115 150 2e-36
YDR148c 99.4 4e-21
SPCC794.07 98.2 9e-21
Hs19923748 97.1 2e-20
YNL071w 94.7 1e-19
7297250 93.2 3e-19
CE14480 92.8 4e-19
SPBC776.15c 92.4 5e-19
Hs4505699 87.8 1e-17
At1g54220 84.3 1e-16
CE09597 84.0 2e-16
7299435 83.6 3e-16
At3g13930 83.2 3e-16
At3g25860 82.4 5e-16
Hs21630255 79.7 4e-15
Hs17458911 79.3 5e-15
CE15651 77.4 2e-14
At1g34430 72.0 8e-13
At5g55070 70.9 2e-12
At3g52200 68.9 6e-12
At4g26910 67.0 2e-11
At5g44950 31.6 0.98
At1g79950 30.8 1.7
YGR227w 30.8 2.0
> 7293036
Length=462
Score = 157 bits (398), Expect = 9e-39, Method: Compositional matrix adjust.
Identities = 83/186 (44%), Positives = 117/186 (62%), Gaps = 9/186 (4%)
Query 15 RISLTAFLIKAISLAINEYPIVNSKFNTETQNSYTEFGSHNVSVAIDSPGGLVVPCVKNV 74
+++ F IKA S+A+++YPIVNS + ++ S G+HN+SVAID+P GLVVP +KN
Sbjct 286 KLTFMPFCIKAASIALSKYPIVNSSLDLASE-SLVFKGAHNISVAIDTPQGLVVPNIKNC 344
Query 75 QDLTLVEIQRELVRLQGLAKANRLSPADLTGGTIALSNVGVISGTYIHPLLFDGQAVIVG 134
Q T++EI ++L L + LSPAD GT +LSN+GVI GTY HP + Q I
Sbjct 345 QTKTIIEIAKDLNALVERGRTGSLSPADFADGTFSLSNIGVIGGTYTHPCIMAPQVAIGA 404
Query 135 VGRVQQLPRFVDSEDSDGKKNTTLEARDIVNCSFSADHRHCDGATITRFSKSIKNLLENP 194
+GR + +PRF D K+ ++A +++ S+SADHR DG T+ FS K LENP
Sbjct 405 MGRTKAVPRFND-------KDEVVKAY-VMSVSWSADHRVIDGVTMASFSNVWKQYLENP 456
Query 195 ALMLVH 200
AL L+H
Sbjct 457 ALFLLH 462
> At3g06850
Length=483
Score = 155 bits (393), Expect = 4e-38, Method: Compositional matrix adjust.
Identities = 82/202 (40%), Positives = 123/202 (60%), Gaps = 9/202 (4%)
Query 1 LRHALNKELASQNIRISLTAFLIKAISLAINEYPIVNSKFNTETQNSYTEFGSHNVSVAI 60
L+ + I+ + LIK++S+A+ +YP VNS FN E+ + GSHN+ VA+
Sbjct 291 LKQFFKENNTDSTIKHTFLPTLIKSLSMALTKYPFVNSCFNAESLEIILK-GSHNIGVAM 349
Query 61 DSPGGLVVPCVKNVQDLTLVEIQRELVRLQGLAKANRLSPADLTGGTIALSNVGVISGTY 120
+ GLVVP +KNVQ L+L+EI +EL RLQ LA N+L+P D+TGGTI LSN+G I G +
Sbjct 350 ATEHGLVVPNIKNVQSLSLLEITKELSRLQHLAANNKLNPEDVTGGTITLSNIGAIGGKF 409
Query 121 IHPLLFDGQAVIVGVGRVQQLPRFVDSEDSDGKKNTTLEARDIVNCSFSADHRHCDGATI 180
PLL + I+ +GR++++P+F K T+ I+ + +ADHR DGAT+
Sbjct 410 GSPLLNLPEVAIIALGRIEKVPKF--------SKEGTVYPASIMMVNIAADHRVLDGATV 461
Query 181 TRFSKSIKNLLENPALMLVHLR 202
RF K +E P L+++ +R
Sbjct 462 ARFCCQWKEYVEKPELLMLQMR 483
> Hs4503265
Length=482
Score = 155 bits (392), Expect = 5e-38, Method: Compositional matrix adjust.
Identities = 84/202 (41%), Positives = 123/202 (60%), Gaps = 9/202 (4%)
Query 1 LRHALNKELASQNIRISLTAFLIKAISLAINEYPIVNSKFNTETQNSYTEFGSHNVSVAI 60
LR L ++ I++S F +KA SL + ++PI+N+ + QN T SHN+ +A+
Sbjct 290 LREELKPIAFARGIKLSFMPFFLKAASLGLLQFPILNASVDENCQN-ITYKASHNIGIAM 348
Query 61 DSPGGLVVPCVKNVQDLTLVEIQRELVRLQGLAKANRLSPADLTGGTIALSNVGVISGTY 120
D+ GL+VP VKNVQ ++ +I EL RLQ L +LS DLTGGT LSN+G I GT+
Sbjct 349 DTEQGLIVPNVKNVQICSIFDIATELNRLQKLGSVGQLSTTDLTGGTFTLSNIGSIGGTF 408
Query 121 IHPLLFDGQAVIVGVGRVQQLPRFVDSEDSDGKKNTTLEARDIVNCSFSADHRHCDGATI 180
P++ + I +G ++ +PRF +K +A+ I+N S+SADHR DGAT+
Sbjct 409 AKPVIMPPEVAIGALGSIKAIPRF-------NQKGEVYKAQ-IMNVSWSADHRVIDGATM 460
Query 181 TRFSKSIKNLLENPALMLVHLR 202
+RFS K+ LENPA ML+ L+
Sbjct 461 SRFSNLWKSYLENPAFMLLDLK 482
> CE01115
Length=448
Score = 150 bits (378), Expect = 2e-36, Method: Compositional matrix adjust.
Identities = 81/197 (41%), Positives = 120/197 (60%), Gaps = 10/197 (5%)
Query 7 KELASQ-NIRISLTAFLIKAISLAINEYPIVNSKFNTETQNSYTEFGSHNVSVAIDSPGG 65
KE A + +I++S F IKA SLA+ EYP +NS + + +N + SHN+ +A+D+PGG
Sbjct 261 KEFAKERHIKLSYMPFFIKAASLALLEYPSLNSTTDEKMENVIHK-ASHNICLAMDTPGG 319
Query 66 LVVPCVKNVQDLTLVEIQRELVRLQGLAKANRLSPADLTGGTIALSNVGVISGTYIHPLL 125
LVVP +KN + ++ EI +EL RL K ++ DL GT +LSN+G I GTY P++
Sbjct 320 LVVPNIKNCEQRSIFEIAQELNRLLEAGKKQQIKREDLIDGTFSLSNIGNIGGTYASPVV 379
Query 126 FDGQAVIVGVGRVQQLPRFVDSEDSDGKKNTTLEARDIVNCSFSADHRHCDGATITRFSK 185
F Q I +G++++LPRF K+ + +I+ S+ ADHR DGAT+ RFS
Sbjct 380 FPPQVAIGAIGKIEKLPRF--------DKHDNVIPVNIMKVSWCADHRVVDGATMARFSN 431
Query 186 SIKNLLENPALMLVHLR 202
K LE+P+ ML L+
Sbjct 432 RWKFYLEHPSAMLAQLK 448
> YDR148c
Length=463
Score = 99.4 bits (246), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 58/185 (31%), Positives = 99/185 (53%), Gaps = 12/185 (6%)
Query 15 RISLTAFLIKAISLAINEYPIVNSKFNTETQNSYTEFGSHNVSVAIDSPGGLVVPCVKNV 74
+ KA +LA + P VN + Q Y ++ ++SVA+ +P GLV P V+N
Sbjct 290 KFGFMGLFSKACTLAAKDIPAVNGAIEGD-QIVYRDY--TDISVAVATPKGLVTPVVRNA 346
Query 75 QDLTLVEIQRELVRLQGLAKANRLSPADLTGGTIALSNVGVISGTYIHPLLFDGQAVIVG 134
+ L++++I+ E+VRL A+ +L+ D+TGGT +SN GV Y P++ Q ++G
Sbjct 347 ESLSVLDIENEIVRLSHKARDGKLTLEDMTGGTFTISNGGVFGSLYGTPIINSPQTAVLG 406
Query 135 VGRVQQLPRFVDSEDSDGKKNTTLEARDIVNCSFSADHRHCDGATITRFSKSIKNLLENP 194
+ V++ P V N + +R ++ + + DHR DG F K++K L+E+P
Sbjct 407 LHGVKERPVTV---------NGQIVSRPMMYLALTYDHRLLDGREAVTFLKTVKELIEDP 457
Query 195 ALMLV 199
ML+
Sbjct 458 RKMLL 462
> SPCC794.07
Length=483
Score = 98.2 bits (243), Expect = 9e-21, Method: Compositional matrix adjust.
Identities = 60/194 (30%), Positives = 97/194 (50%), Gaps = 10/194 (5%)
Query 1 LRHALNKELASQNIRISLTAFLIKAISLAINEYPIVNSKFNTETQNSYTEFGSHNVSVAI 60
LR ALN +A ++S+ +IKA + A+ + P VN+ + + Y ++S+A+
Sbjct 295 LRAALNA-MADGRYKLSVNDLVIKATTAALRQVPEVNAAWMGDFIRQYKNV---DISMAV 350
Query 61 DSPGGLVVPCVKNVQDLTLVEIQRELVRLQGLAKANRLSPADLTGGTIALSNVGVISGTY 120
+P GL+ P ++N L L EI A+ N+L P + GGT +SN+G+
Sbjct 351 ATPSGLITPVIRNTHALGLAEISTLAKDYGQRARNNKLKPEEYQGGTFTISNLGMFPVDQ 410
Query 121 IHPLLFDGQAVIVGVGRVQQLPRFVDSEDSDGKKNTTLEARDIVNCSFSADHRHCDGATI 180
++ QA I+ VG VD+ D + I+ C+ S+DHR DGA
Sbjct 411 FTAIINPPQACILAVGTT------VDTVVPDSTSEKGFKVAPIMKCTLSSDHRVVDGAMA 464
Query 181 TRFSKSIKNLLENP 194
RF+ ++K +LENP
Sbjct 465 ARFTTALKKILENP 478
> Hs19923748
Length=453
Score = 97.1 bits (240), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 58/202 (28%), Positives = 107/202 (52%), Gaps = 14/202 (6%)
Query 3 HALNKE--LASQNIRISLTAFLIKAISLAINEYPIVNSKFNTETQNS-YTEFGSHNVSVA 59
A +KE L N+++ + +KA + A+ E P+VN+ + T+ Y ++ ++SVA
Sbjct 263 RARHKEAFLKKHNLKLGFMSAFVKASAFALQEQPVVNAVIDDTTKEVVYRDY--IDISVA 320
Query 60 IDSPGGLVVPCVKNVQDLTLVEIQRELVRLQGLAKANRLSPADLTGGTIALSNVGVISGT 119
+ +P GLVVP ++NV+ + +I+R + L A+ N L+ D+ GGT +SN GV
Sbjct 321 VATPRGLVVPVIRNVEAMNFADIERTITELGEKARKNELAIEDMDGGTFTISNGGVFGSL 380
Query 120 YIHPLLFDGQAVIVGVGRVQQLPRFVDSEDSDGKKNTTLEARDIVNCSFSADHRHCDGAT 179
+ P++ Q+ I+G+ + P + + +E R ++ + + DHR DG
Sbjct 381 FGTPIINPPQSAILGMHGIFDRPVAIGGK---------VEVRPMMYVALTYDHRLIDGRE 431
Query 180 ITRFSKSIKNLLENPALMLVHL 201
F + IK +E+P ++L+ L
Sbjct 432 AVTFLRKIKAAVEDPRVLLLDL 453
> YNL071w
Length=482
Score = 94.7 bits (234), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 62/200 (31%), Positives = 109/200 (54%), Gaps = 9/200 (4%)
Query 1 LRHALNKELASQNIRISLTAFLIKAISLAINEYPIVNSKFNTETQNSYTEFGSHNVSVAI 60
LR +LN A+ ++S+ L+KAI++A P N+ + +N +F + +VSVA+
Sbjct 291 LRQSLNA-TANDKYKLSINDLLVKAITVAAKRVPDANA-YWLPNENVIRKFKNVDVSVAV 348
Query 61 DSPGGLVVPCVKNVQDLTLVEIQRELVRLQGLAKANRLSPADLTGGTIALSNVGVISGTY 120
+P GL+ P VKN + L +I E+ L A+ N+L+P + GGTI +SN+G+ +
Sbjct 349 ATPTGLLTPIVKNCEAKGLSQISNEIKELVKRARINKLAPEEFQGGTICISNMGMNNAVN 408
Query 121 IHPLLFD-GQAVIVGVGRVQQLPRFVDSEDSDGKKNTTLEARDIVNCSFSADHRHCDGAT 179
+ + + Q+ I+ + V++ V ED+ + + + + + +F DHR DGA
Sbjct 409 MFTSIINPPQSTILAIATVER----VAVEDAAAENGFSFDNQVTITGTF--DHRTIDGAK 462
Query 180 ITRFSKSIKNLLENPALMLV 199
F K +K ++ENP ML+
Sbjct 463 GAEFMKELKTVIENPLEMLL 482
> 7297250
Length=512
Score = 93.2 bits (230), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 58/200 (29%), Positives = 105/200 (52%), Gaps = 11/200 (5%)
Query 1 LRHALNKELASQNIRISLTAFLIKAISLAINEYPIVNSKFNTETQNSYTEFGSHNVSVAI 60
R +NK+ Q R+S+ F+IKA+++A + P NS + Y + +VSVA+
Sbjct 323 FRAKVNKKYEKQGARVSVNDFIIKAVAIASLKVPEANSAWMDTVIRKYDDV---DVSVAV 379
Query 61 DSPGGLVVPCVKNVQDLTLVEIQRELVRLQGLAKANRLSPADLTGGTIALSNVGVISGTY 120
+ GL+ P V N ++EI +++ L A+ N+L P + GGTI++SN+G+
Sbjct 380 STDKGLITPIVFNADRKGVLEISKDVKALAAKARDNKLQPHEFQGGTISVSNLGMFGVNQ 439
Query 121 IHPLLFDGQAVIVGVG-RVQQLPRFVDSEDSDGKKNTTLEARDIVNCSFSADHRHCDGAT 179
++ Q+ I+ +G +QL D + G K +++ + SADHR DGA
Sbjct 440 FAAVINPPQSCILAIGTTTKQL--VADPDSLKGFKEV-----NMLTVTLSADHRVVDGAV 492
Query 180 ITRFSKSIKNLLENPALMLV 199
R+ + ++ +E+P+ M++
Sbjct 493 AARWLQHFRDYMEDPSNMVL 512
> CE14480
Length=452
Score = 92.8 bits (229), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 57/202 (28%), Positives = 102/202 (50%), Gaps = 24/202 (11%)
Query 1 LRHALNKE-LASQNIRISLTAFLIKAISLAINEYPIVNSKFNTETQNSYTEFGSHNVSVA 59
+R K+ +A +++ + + ++A + A+ E P+VN+ + E + Y F ++SVA
Sbjct 274 MRKTYQKDFVAKHGVKLGMMSPFVRAAAYALQESPVVNAVLD-ENEIVYRHF--VDISVA 330
Query 60 IDSPGGLVVPCVKNVQDLTLVEIQRELVRLQGLAKANRLSPADLTGGTIALSNVGVISGT 119
+ +P GLVVP ++NV+ + +I+ EL L A+ +L+ D+ GGT +SN GV
Sbjct 331 VATPKGLVVPVLRNVESMNYAQIELELANLGVKARDGKLAVEDMEGGTFTISNGGVFGSM 390
Query 120 YIHPLLFDGQAVIVGVGRVQQLPRFVDSEDSDGKKNTTLEARDIVNCSFSADHRHCDGAT 179
+ P++ Q+ I+G+ E R I+ + + DHR DG
Sbjct 391 FGTPIINPPQSAILGM--------------------HGPEIRPIMQIALTYDHRLIDGRE 430
Query 180 ITRFSKSIKNLLENPALMLVHL 201
F K IK +E+P +M ++L
Sbjct 431 AVTFLKKIKTAVEDPRIMFMNL 452
> SPBC776.15c
Length=452
Score = 92.4 bits (228), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 54/203 (26%), Positives = 101/203 (49%), Gaps = 15/203 (7%)
Query 1 LRHALNKE-LASQNIRISLTAFLIKAISLAINEYPIVNSKFNTETQNS---YTEFGSHNV 56
LR E L ++I +F KA + A+ + P +N E + Y +F ++
Sbjct 260 LRKKYKDEILKETGVKIGFMSFFSKACTQAMKQIPAINGSIEGEGKGDTLVYRDF--CDL 317
Query 57 SVAIDSPGGLVVPCVKNVQDLTLVEIQRELVRLQGLAKANRLSPADLTGGTIALSNVGVI 116
S+A+ +P GLV P ++N + ++L+EI+ + L A+A +L+ D+ GT +SN G+
Sbjct 318 SIAVATPKGLVTPVIRNAESMSLLEIESAIATLGSKARAGKLAIEDMASGTFTISNGGIF 377
Query 117 SGTYIHPLLFDGQAVIVGVGRVQQLPRFVDSEDSDGKKNTTLEARDIVNCSFSADHRHCD 176
Y P++ Q ++G+ +++ P + N + R ++ + + DHR D
Sbjct 378 GSLYGTPIINLPQTAVLGLHAIKERPVVI---------NGQVVPRPMMYLALTYDHRMVD 428
Query 177 GATITRFSKSIKNLLENPALMLV 199
G F + +K +E+PA ML+
Sbjct 429 GREAVTFLRLVKEYIEDPAKMLL 451
> Hs4505699
Length=501
Score = 87.8 bits (216), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 55/190 (28%), Positives = 99/190 (52%), Gaps = 7/190 (3%)
Query 5 LNKELASQNIRISLTAFLIKAISLAINEYPIVNSKFNTETQNSYTEFGSHNVSVAIDSPG 64
+ ++L +I++S+ F+IKA ++ + + P VN ++ E ++SVA+ +
Sbjct 315 VRQDLVKDDIKVSVNDFIIKAAAVTLKQMPDVNVSWDGEGPKQLPFI---DISVAVATDK 371
Query 65 GLVVPCVKNVQDLTLVEIQRELVRLQGLAKANRLSPADLTGGTIALSNVGVISGTYIHPL 124
GL+ P +K+ + EI + L A+ +L P + GG+ ++SN+G+ +
Sbjct 372 GLLTPIIKDAAAKGIQEIADSVKALSKKARDGKLLPEEYQGGSFSISNLGMFGIDEFTAV 431
Query 125 LFDGQAVIVGVGRVQQLPRFVDSEDSDGKKNTTLEARDIVNCSFSADHRHCDGATITRFS 184
+ QA I+ VGR + + + +ED +G N L+ R ++ + S+D R D TRF
Sbjct 432 INPPQACILAVGRFRPVLKL--TEDEEG--NAKLQQRQLITVTMSSDSRVVDDELATRFL 487
Query 185 KSIKNLLENP 194
KS K LENP
Sbjct 488 KSFKANLENP 497
> At1g54220
Length=424
Score = 84.3 bits (207), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 66/201 (32%), Positives = 96/201 (47%), Gaps = 12/201 (5%)
Query 1 LRHALNK-ELASQNIRISLTAFLIKAISLAINEYPIVNSKFNTETQNSYTEFGSHNVSVA 59
LR LN + AS RIS+ ++KA +LA+ + P NS + T + +F + N++VA
Sbjct 234 LRSQLNSFKEASGGKRISVNDLVVKAAALALRKVPQCNSSW---TDDYIRQFKNVNINVA 290
Query 60 IDSPGGLVVPCVKNVQDLTLVEIQRELVRLQGLAKANRLSPADLTGGTIALSNVGVISG- 118
+ + GL VP VK+ L I E+ L AK N L P D GGT +SN+G G
Sbjct 291 VQTENGLYVPVVKDADRKGLSTIGEEVRLLAQKAKENSLKPEDYEGGTFTVSNLGGPFGI 350
Query 119 TYIHPLLFDGQAVIVGVGRVQQLPRFVDSEDSDGKKNTTLEARDIVNCSFSADHRHCDGA 178
++ QA I+ VG ++ R V D + + S DHR DGA
Sbjct 351 KQFCAVVNPPQAAILAVGSAEK--RVVPGNGPD-----QFNFASYMPVTLSCDHRVVDGA 403
Query 179 TITRFSKSIKNLLENPALMLV 199
+ K+ K +ENP ML+
Sbjct 404 IGAEWLKAFKGYIENPKSMLL 424
> CE09597
Length=507
Score = 84.0 bits (206), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 61/197 (30%), Positives = 94/197 (47%), Gaps = 14/197 (7%)
Query 5 LNKELASQNIRISLTAFLIKAISLAINEYPIVNSKFNTETQNSYTEFGSH-NVSVAIDSP 63
L K + Q +IS+ F+IKA +LA P NS + +S+ H +VSVA+ +P
Sbjct 323 LAKGTSGQATKISINDFIIKASALACQRVPEANSYW----MDSFIRENHHVDVSVAVSTP 378
Query 64 GGLVVPCVKNVQDLTLVEIQRELVRLQGLAKANRLSPADLTGGTIALSNVGVI-SGTYIH 122
GL+ P + N L I E+V L A+ +L P + GGT +SN+G+ S +
Sbjct 379 AGLITPIIFNAHAKGLATIASEIVELAQRAREGKLQPHEFQGGTFTVSNLGMFGSVSDFT 438
Query 123 PLLFDGQAVIVGVGRVQQLPRFVDSEDSDGKKNTTLEARDIVNCSFSADHRHCDGATITR 182
++ Q+ I+ +G + V E KK T++ + S DHR DGA
Sbjct 439 AIINPPQSCILAIGGASD--KLVPDEAEGYKKIKTMK------VTLSCDHRTVDGAVGAV 490
Query 183 FSKSIKNLLENPALMLV 199
+ + K LE P ML+
Sbjct 491 WLRHFKEFLEKPHTMLL 507
> 7299435
Length=468
Score = 83.6 bits (205), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 53/188 (28%), Positives = 97/188 (51%), Gaps = 12/188 (6%)
Query 14 IRISLTAFLIKAISLAINEYPIVNSKFNTETQNSYTEFGSHNVSVAIDSPGGLVVPCVKN 73
I+ + KA + A+ + P+VN+ + T Y ++ ++SVA+ +P GLVVP ++N
Sbjct 293 IKFGFMSIFAKASAYALQDQPVVNAVIDG-TDIVYRDY--VDISVAVATPRGLVVPVIRN 349
Query 74 VQDLTLVEIQRELVRLQGLAKANRLSPADLTGGTIALSNVGVISGTYIHPLLFDGQAVIV 133
V+ + +I+ L L A+ + ++ D+ GGT +SN GV P++ Q+ I+
Sbjct 350 VEGMNYADIEIALAGLADKARRDAITVEDMDGGTFTISNGGVFGSLMGTPIINPPQSAIL 409
Query 134 GVGRVQQLPRFVDSEDSDGKKNTTLEARDIVNCSFSADHRHCDGATITRFSKSIKNLLEN 193
G+ + + P V E ++ R ++ + + DHR DG F + IK +EN
Sbjct 410 GMHGIFERPIAVKGE---------VKIRPMMYIALTYDHRIIDGREAVLFLRKIKAAVEN 460
Query 194 PALMLVHL 201
PA+++ L
Sbjct 461 PAIIVAGL 468
> At3g13930
Length=508
Score = 83.2 bits (204), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 67/205 (32%), Positives = 96/205 (46%), Gaps = 20/205 (9%)
Query 1 LRHALNK-ELASQNIRISLTAFLIKAISLAINEYPIVNSKFNTETQNSYTEFGSHNVSVA 59
LR LN + AS RIS+ +IKA +LA+ + P NS + T +F + N++VA
Sbjct 318 LRSQLNSFQEASGGKRISVNDLVIKAAALALRKVPQCNSSW---TDEYIRQFKNVNINVA 374
Query 60 IDSPGGLVVPCVKNVQDLTLVEIQRELVRLQGLAKANRLSPADLTGGTIALSNVGVISG- 118
+ + GL VP VK+ L I E+ L AK N L P D GGT +SN+G G
Sbjct 375 VQTENGLYVPVVKDADKKGLSTIGEEVRFLAQKAKENSLKPEDYEGGTFTVSNLGGPFGI 434
Query 119 ----TYIHPLLFDGQAVIVGVGRVQQLPRFVDSEDSDGKKNTTLEARDIVNCSFSADHRH 174
I+P QA I+ +G ++ R V D ++ + S DHR
Sbjct 435 KQFCAVINP----PQAAILAIGSAEK--RVVPGTGPD-----QYNVASYMSVTLSCDHRV 483
Query 175 CDGATITRFSKSIKNLLENPALMLV 199
DGA + K+ K +E P ML+
Sbjct 484 IDGAIGAEWLKAFKGYIETPESMLL 508
> At3g25860
Length=480
Score = 82.4 bits (202), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 59/182 (32%), Positives = 93/182 (51%), Gaps = 16/182 (8%)
Query 16 ISLTAFLIKAISLAINEYPIVNSKFNTETQNSYTEFGSHNVSVAIDSPGGLVVPCVKNVQ 75
+++TA L KA +A+ ++P+VN+ SY S N++VA+ GGL+ P +++
Sbjct 307 VTMTALLAKAAGMALAQHPVVNASCKDGKSFSYNS--SINIAVAVAINGGLITPVLQDAD 364
Query 76 DLTLVEIQRELVRLQGLAKANRLSPADLTGGTIALSNVGVISGTYIHPLLFDGQAVIVGV 135
L L + ++ L G A++ +L P + GT LSN+G+ +L GQ I+ V
Sbjct 365 KLDLYLLSQKWKELVGKARSKQLQPHEYNSGTFTLSNLGMFGVDRFDAILPPGQGAIMAV 424
Query 136 GRVQQLPRFVDSEDSDG---KKNTTLEARDIVNCSFSADHRHCDGATITRFSKSIKNLLE 192
G + P V D DG KNT L VN +ADHR GA + F ++ ++E
Sbjct 425 GASK--PTVV--ADKDGFFSVKNTML-----VNV--TADHRIVYGADLAAFLQTFAKIIE 473
Query 193 NP 194
NP
Sbjct 474 NP 475
> Hs21630255
Length=613
Score = 79.7 bits (195), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 58/201 (28%), Positives = 98/201 (48%), Gaps = 15/201 (7%)
Query 1 LRHALNKELASQNIRISLTAFLIKAISLAINEYPIVNSKFNTET--QNSYTEFGSHNVSV 58
+R LNK L ++ +IS+ F+IKA +LA + P NS + QN +VSV
Sbjct 426 VRKELNKILEGRS-KISVNDFIIKASALACLKVPEANSSWMDTVIRQNHVV-----DVSV 479
Query 59 AIDSPGGLVVPCVKNVQDLTLVEIQRELVRLQGLAKANRLSPADLTGGTIALSNVGVISG 118
A+ +P GL+ P V N + I ++V L A+ +L P + GGT +SN+G+
Sbjct 480 AVSTPAGLITPIVFNAHIKGVETIANDVVSLATKAREGKLQPHEFQGGTFTISNLGMFGI 539
Query 119 TYIHPLLFDGQAVIVGVGRVQQLPRFVDSEDSDGKKNTTLEARDIVNCSFSADHRHCDGA 178
++ QA I+ +G + + V +++ G + +++ + S DHR DGA
Sbjct 540 KNFSAIINPPQACILAIGASED--KLVPADNEKG-----FDVASMMSVTLSCDHRVVDGA 592
Query 179 TITRFSKSIKNLLENPALMLV 199
++ + LE P ML+
Sbjct 593 VGAQWLAEFRKYLEKPITMLL 613
> Hs17458911
Length=647
Score = 79.3 bits (194), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 57/200 (28%), Positives = 101/200 (50%), Gaps = 13/200 (6%)
Query 1 LRHALNKELASQNIRISLTAFLIKAISLAINEYPIVNSKF-NTETQNSYTEFGSHNVSVA 59
+R LNK L ++ +IS+ F+IKA +LA + P NS + +T + ++ +VSVA
Sbjct 460 VRKELNKILEGRS-KISVNDFIIKASALACLKVPEANSSWMDTVIRQNHV----VDVSVA 514
Query 60 IDSPGGLVVPCVKNVQDLTLVEIQRELVRLQGLAKANRLSPADLTGGTIALSNVGVISGT 119
+ +P GL+ P V N + I ++V L A+ +L P + GGT +SN+G+
Sbjct 515 VSTPAGLITPIVFNAHIKGVETIANDVVSLATKAREGKLQPHEFQGGTFTISNLGMFGIK 574
Query 120 YIHPLLFDGQAVIVGVGRVQQLPRFVDSEDSDGKKNTTLEARDIVNCSFSADHRHCDGAT 179
++ QA I+ +G + + V +++ G + +++ + S DHR DGA
Sbjct 575 NFSAIINPPQACILAIGASED--KLVPADNEKG-----FDVASMMSVTLSCDHRVVDGAV 627
Query 180 ITRFSKSIKNLLENPALMLV 199
++ + LE P ML+
Sbjct 628 GAQWLAEFRKYLEKPITMLL 647
> CE15651
Length=337
Score = 77.4 bits (189), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 55/198 (27%), Positives = 93/198 (46%), Gaps = 14/198 (7%)
Query 4 ALNKELASQNIRISLTAFLIKAISLAINEYPIVNSKFNTETQNSYTEFGSHNVSVAIDSP 63
AL ++L +SL F+IKA +LA+ P VN ++ E GS ++SVA+ +P
Sbjct 139 ALRQKLKKSGTAVSLNDFIIKAAALALRSVPTVNVRWTPEG----IGLGSVDISVAVATP 194
Query 64 GGLVVPCVKNVQDLTLVEIQRELVRLQGLAKANRLSPADLTGGTIALSNVGVI-SGTYIH 122
GL+ P V+N L ++ I ++ L GLA+ ++L P GG+ +SN+G+ S T
Sbjct 195 TGLITPIVENSDILGVLAISSKVKELSGLARESKLKPQQFQGGSFTISNLGMFGSVTNFT 254
Query 123 PLLFDGQAVIVGVGRVQQLPRFVDSEDSDGKKNTTLEARDIVNCSFSADHRHCDGATITR 182
++ Q I+ +G + VD + LE + ++ + D R R
Sbjct 255 AIINPPQCAILTIGGTRSEVVSVDGQ---------LETQKLMGVNLCFDGRAISEECAKR 305
Query 183 FSKSIKNLLENPALMLVH 200
F L +P L++
Sbjct 306 FLLHFSESLSDPELLIAE 323
> At1g34430
Length=465
Score = 72.0 bits (175), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 49/163 (30%), Positives = 83/163 (50%), Gaps = 12/163 (7%)
Query 33 YPIVNSKFNTETQNSYTEFGSHNVSVAIDSPGGLVVPCVKNVQDLTLVEIQRELVRLQGL 92
+P+VNS + NS+ S NV+VA+ GGL+ P ++N + + + R+ L
Sbjct 309 HPVVNS--SCRDGNSFVYNSSINVAVAVAIDGGLITPVLQNADKVDIYSLSRKWKELVDK 366
Query 93 AKANRLSPADLTGGTIALSNVGVISGTYIHPLLFDGQAVIVGVGRVQQLPRFVDSEDSD- 151
A+A +L P + GT LSN+G+ +L G I+ VG Q P V ++D
Sbjct 367 ARAKQLQPQEYNTGTFTLSNLGMFGVDRFDAILPPGTGAIMAVGASQ--PSVVATKDGRI 424
Query 152 GKKNTTLEARDIVNCSFSADHRHCDGATITRFSKSIKNLLENP 194
G KN + + +ADHR GA + +F +++ +++E+P
Sbjct 425 GMKNQ-------MQVNVTADHRVIYGADLAQFLQTLASIIEDP 460
> At5g55070
Length=464
Score = 70.9 bits (172), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 46/193 (23%), Positives = 95/193 (49%), Gaps = 12/193 (6%)
Query 9 LASQNIRISLTAFLIKAISLAINEYPIVNSKFNTETQNSYTEFGSHNVSVAIDSPGGLVV 68
L +++ L + IKA A+ P+VN+ + + Y ++ ++S+A+ + GLVV
Sbjct 284 LEKHGVKLGLMSGFIKAAVSALQHQPVVNAVIDGD-DIIYRDY--VDISIAVGTSKGLVV 340
Query 69 PCVKNVQDLTLVEIQRELVRLQGLAKANRLSPADLTGGTIALSNVGVISGTYIHPLLFDG 128
P +++ + +I++ + L A +S ++ GG+ +SN GV P++
Sbjct 341 PVIRDADKMNFADIEKTINGLAKKATEGTISIDEMAGGSFTVSNGGVYGSLISTPIINPP 400
Query 129 QAVIVGVGRVQQLPRFVDSEDSDGKKNTTLEARDIVNCSFSADHRHCDGATITRFSKSIK 188
Q+ I+G+ + Q P V ++ R ++ + + DHR DG F + IK
Sbjct 401 QSAILGMHSIVQRPMVV---------GGSVVPRPMMYVALTYDHRLIDGREAVYFLRRIK 451
Query 189 NLLENPALMLVHL 201
+++E+P +L+ +
Sbjct 452 DVVEDPQRLLLDI 464
> At3g52200
Length=637
Score = 68.9 bits (167), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 50/191 (26%), Positives = 96/191 (50%), Gaps = 6/191 (3%)
Query 4 ALNKEL-ASQNIRISLTAFLIKAISLAINEYPIVNSKFNTETQNSYTEFGSHNVSVAIDS 62
A KEL + +++S+ +IKA+++A+ N+ ++ E + S ++S+A+ +
Sbjct 446 AFRKELQENHGVKVSVNDIVIKAVAVALRNVRQANAFWDAE-KGDIVMCDSVDISIAVAT 504
Query 63 PGGLVVPCVKNVQDLTLVEIQRELVRLQGLAKANRLSPADLTGGTIALSNVGVISGTYIH 122
GL+ P +KN ++ I E+ L A++ +L+P + GGT ++SN+G+
Sbjct 505 EKGLMTPIIKNADQKSISAISLEVKELAQKARSGKLAPHEFQGGTFSISNLGMYPVDNFC 564
Query 123 PLLFDGQAVIVGVGRVQQLPRFVDSEDSDGKKNTTLEARDIVNCSFSADHRHCDGATITR 182
++ QA I+ VGR ++ V D K + + +N + SADHR DG
Sbjct 565 AIINPPQAGILAVGRGNKVVEPVIGLDGIEKPSVVTK----MNVTLSADHRIFDGQVGAS 620
Query 183 FSKSIKNLLEN 193
F +++ E+
Sbjct 621 FMSELRSNFED 631
> At4g26910
Length=511
Score = 67.0 bits (162), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 44/188 (23%), Positives = 92/188 (48%), Gaps = 12/188 (6%)
Query 14 IRISLTAFLIKAISLAINEYPIVNSKFNTETQNSYTEFGSHNVSVAIDSPGGLVVPCVKN 73
+++ L + IKA A+ P+VN+ + + Y ++ ++S+A+ + GLVVP ++
Sbjct 336 VKLGLMSGFIKAAVSALQHQPVVNAVIDGD-DIIYRDY--VDISIAVGTSKGLVVPVIRG 392
Query 74 VQDLTLVEIQRELVRLQGLAKANRLSPADLTGGTIALSNVGVISGTYIHPLLFDGQAVIV 133
+ EI++ + L A +S ++ GG+ +SN GV P++ Q+ I+
Sbjct 393 ADKMNFAEIEKTINSLAKKANEGTISIDEMAGGSFTVSNGGVYGSLISTPIINPPQSAIL 452
Query 134 GVGRVQQLPRFVDSEDSDGKKNTTLEARDIVNCSFSADHRHCDGATITRFSKSIKNLLEN 193
G+ + P V ++ R ++ + + DHR DG F + +K+++E+
Sbjct 453 GMHSIVSRPMVVGG---------SVVPRPMMYVALTYDHRLIDGREAVYFLRRVKDVVED 503
Query 194 PALMLVHL 201
P +L+ +
Sbjct 504 PQRLLLDI 511
> At5g44950
Length=438
Score = 31.6 bits (70), Expect = 0.98, Method: Compositional matrix adjust.
Identities = 31/106 (29%), Positives = 46/106 (43%), Gaps = 19/106 (17%)
Query 52 GSHNVSVAIDSPGGLVVP---------CVKNVQDLTLVEIQRELVRLQGLAKANRLSPAD 102
GS++ SV ID+PG + VKN+ L +++ E GL L P D
Sbjct 215 GSNDFSVEIDAPGLKYMSFRDSQSDRIVVKNLSSLVKIDLDTEFNLKFGLGSP--LEPED 272
Query 103 LTGGTIA---LSNVGVISGTYI-HPLLFDGQAVIVGVGRVQQLPRF 144
LT I L+ + + I HP L V+ ++ QLP+F
Sbjct 273 LTKRDIIRDFLTGISSVKHMIISHPTL----EVLYRYSKIGQLPKF 314
> At1g79950
Length=983
Score = 30.8 bits (68), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 20/84 (23%), Positives = 36/84 (42%), Gaps = 0/84 (0%)
Query 83 QRELVRLQGLAKANRLSPADLTGGTIALSNVGVISGTYIHPLLFDGQAVIVGVGRVQQLP 142
Q EL RL+ ANR SP + G + + + ++ T P + G + R +L
Sbjct 760 QNELSRLEAFPPANRASPLERDGNNVKRNGLTILKHTGKIPRIVKGDVMQGCSSRKAKLV 819
Query 143 RFVDSEDSDGKKNTTLEARDIVNC 166
D E++ ++ + + NC
Sbjct 820 ELSDDEETPVERRCEVVDLESDNC 843
> YGR227w
Length=525
Score = 30.8 bits (68), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 18/46 (39%), Positives = 25/46 (54%), Gaps = 4/46 (8%)
Query 28 LAINEYPIVNSKFNTETQNSYTEFGSHNVSVAIDSPGGLVVPCVKN 73
LA+ I+ +FNT T N+Y + H AID LV+P +KN
Sbjct 226 LAVERPAILQKQFNTHTFNNYLKLFIH----AIDDFSNLVLPYMKN 267
Lambda K H
0.319 0.134 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3551102874
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40