bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0898_orf1
Length=120
Score E
Sequences producing significant alignments: (Bits) Value
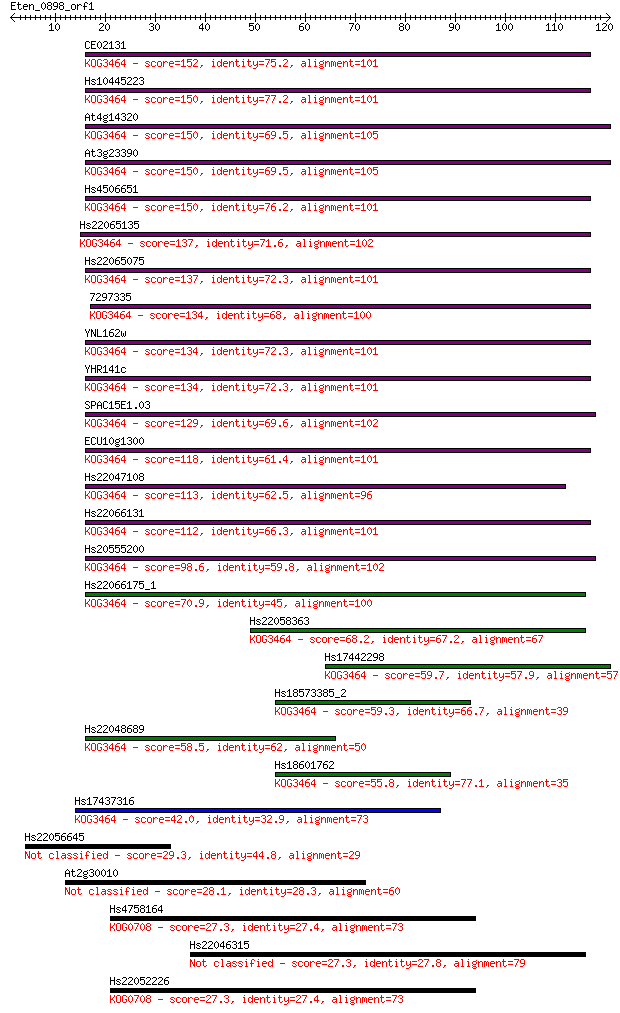
CE02131 152 2e-37
Hs10445223 150 4e-37
At4g14320 150 5e-37
At3g23390 150 5e-37
Hs4506651 150 6e-37
Hs22065135 137 3e-33
Hs22065075 137 3e-33
7297335 134 4e-32
YNL162w 134 6e-32
YHR141c 134 6e-32
SPAC15E1.03 129 1e-30
ECU10g1300 118 2e-27
Hs22047108 113 7e-26
Hs22066131 112 2e-25
Hs20555200 98.6 2e-21
Hs22066175_1 70.9 6e-13
Hs22058363 68.2 3e-12
Hs17442298 59.7 1e-09
Hs18573385_2 59.3 2e-09
Hs22048689 58.5 3e-09
Hs18601762 55.8 2e-08
Hs17437316 42.0 3e-04
Hs22056645 29.3 1.9
At2g30010 28.1 4.5
Hs4758164 27.3 6.6
Hs22046315 27.3 6.6
Hs22052226 27.3 7.1
> CE02131
Length=105
Score = 152 bits (383), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 76/101 (75%), Positives = 86/101 (85%), Gaps = 1/101 (0%)
Query 16 MVNIPKTRNTFCRGSKCRKHTPHKVSQYKKGKDSLVVQGKRRYDAKQKGFGGQTKPIFRK 75
MVN+PK R TFC G KCRKHT HKV+QYKKGK+S QG+RRYD KQ GFGGQTKPIFRK
Sbjct 1 MVNVPKARRTFCDG-KCRKHTNHKVTQYKKGKESKFAQGRRRYDRKQSGFGGQTKPIFRK 59
Query 76 KAKTTKKIVLKLECTKCKTKRLLPIKRCKSFELGADKKAKG 116
KAKTTKKIVL++ECT+CK K+ LPIKRCK FELG KK++G
Sbjct 60 KAKTTKKIVLRMECTECKHKKQLPIKRCKHFELGGQKKSRG 100
> Hs10445223
Length=106
Score = 150 bits (380), Expect = 4e-37, Method: Compositional matrix adjust.
Identities = 78/103 (75%), Positives = 87/103 (84%), Gaps = 4/103 (3%)
Query 16 MVNIPKTRNTFCRGSKCRKHTPHKVSQYKKGKDSLVVQGKRRYDAKQKGFGGQTKPIFRK 75
MVN+PKTR TFC+ KC KH PHKV+QYKKGKDSL QGKRRYD KQ G+GGQTKPIFRK
Sbjct 1 MVNVPKTRRTFCK--KCGKHQPHKVTQYKKGKDSLYAQGKRRYDRKQSGYGGQTKPIFRK 58
Query 76 KAKTTKKIVLKLECTK--CKTKRLLPIKRCKSFELGADKKAKG 116
KAKTTKKIVL+LEC + C++KR+L IKRCK FELG DKK KG
Sbjct 59 KAKTTKKIVLRLECVEPNCRSKRMLAIKRCKHFELGGDKKRKG 101
> At4g14320
Length=105
Score = 150 bits (379), Expect = 5e-37, Method: Compositional matrix adjust.
Identities = 73/105 (69%), Positives = 84/105 (80%), Gaps = 0/105 (0%)
Query 16 MVNIPKTRNTFCRGSKCRKHTPHKVSQYKKGKDSLVVQGKRRYDAKQKGFGGQTKPIFRK 75
MVNIPKT+NT+C+ +C+KHT HKV+QYKKGKDSL QGKRRYD KQ G+GGQTKP+F K
Sbjct 1 MVNIPKTKNTYCKNKECKKHTLHKVTQYKKGKDSLAAQGKRRYDRKQSGYGGQTKPVFHK 60
Query 76 KAKTTKKIVLKLECTKCKTKRLLPIKRCKSFELGADKKAKGGPQF 120
KAKTTKKIVL+L+C CK PIKRCK FE+G DKK KG F
Sbjct 61 KAKTTKKIVLRLQCQSCKHFSQRPIKRCKHFEIGGDKKGKGTSLF 105
> At3g23390
Length=105
Score = 150 bits (379), Expect = 5e-37, Method: Compositional matrix adjust.
Identities = 73/105 (69%), Positives = 84/105 (80%), Gaps = 0/105 (0%)
Query 16 MVNIPKTRNTFCRGSKCRKHTPHKVSQYKKGKDSLVVQGKRRYDAKQKGFGGQTKPIFRK 75
MVNIPKT+NT+C+ +C+KHT HKV+QYKKGKDSL QGKRRYD KQ G+GGQTKP+F K
Sbjct 1 MVNIPKTKNTYCKNKECKKHTLHKVTQYKKGKDSLAAQGKRRYDRKQSGYGGQTKPVFHK 60
Query 76 KAKTTKKIVLKLECTKCKTKRLLPIKRCKSFELGADKKAKGGPQF 120
KAKTTKKIVL+L+C CK PIKRCK FE+G DKK KG F
Sbjct 61 KAKTTKKIVLRLQCQSCKHFSQRPIKRCKHFEIGGDKKGKGTSLF 105
> Hs4506651
Length=106
Score = 150 bits (378), Expect = 6e-37, Method: Compositional matrix adjust.
Identities = 77/103 (74%), Positives = 87/103 (84%), Gaps = 4/103 (3%)
Query 16 MVNIPKTRNTFCRGSKCRKHTPHKVSQYKKGKDSLVVQGKRRYDAKQKGFGGQTKPIFRK 75
MVN+PKTR TFC+ KC KH PHKV+QYKKGKDSL QG+RRYD KQ G+GGQTKPIFRK
Sbjct 1 MVNVPKTRRTFCK--KCGKHQPHKVTQYKKGKDSLYAQGRRRYDRKQSGYGGQTKPIFRK 58
Query 76 KAKTTKKIVLKLECTK--CKTKRLLPIKRCKSFELGADKKAKG 116
KAKTTKKIVL+LEC + C++KR+L IKRCK FELG DKK KG
Sbjct 59 KAKTTKKIVLRLECVEPNCRSKRMLAIKRCKHFELGGDKKRKG 101
> Hs22065135
Length=321
Score = 137 bits (346), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 73/104 (70%), Positives = 85/104 (81%), Gaps = 4/104 (3%)
Query 15 KMVNIPKTRNTFCRGSKCRKHTPHKVSQYKKGKDSLVVQGKRRYDAKQKGFGGQTKPIFR 74
K+VN+PKTR TFC+ KC KH PHKV+QYKKGKDSL QGKRRYD KQ G+G QTKPIF+
Sbjct 215 KVVNVPKTRITFCK--KCGKHQPHKVTQYKKGKDSLYAQGKRRYDQKQSGYGRQTKPIFQ 272
Query 75 KKAKTTKKIVLKLEC--TKCKTKRLLPIKRCKSFELGADKKAKG 116
KKAKTTKKIVL+LEC + C+ +R+L IKR K F+LG DKK KG
Sbjct 273 KKAKTTKKIVLRLECVESNCRFRRMLTIKRYKHFKLGGDKKRKG 316
> Hs22065075
Length=118
Score = 137 bits (346), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 73/103 (70%), Positives = 81/103 (78%), Gaps = 4/103 (3%)
Query 16 MVNIPKTRNTFCRGSKCRKHTPHKVSQYKKGKDSLVVQGKRRYDAKQKGFGGQTKPIFRK 75
MV +PKTR TFC+ KC KH PHKV QYK GKDSL QGK RYD KQ G+GGQTKPIFRK
Sbjct 1 MVKVPKTRRTFCK--KCDKHQPHKVRQYKDGKDSLYAQGKWRYDRKQSGYGGQTKPIFRK 58
Query 76 KAKTTKKIVLKLECTK--CKTKRLLPIKRCKSFELGADKKAKG 116
KAKTTKKIVL LEC + C++KR+L +RCK FELG DKK KG
Sbjct 59 KAKTTKKIVLGLECIQPNCRSKRMLATERCKHFELGGDKKRKG 101
> 7297335
Length=122
Score = 134 bits (337), Expect = 4e-32, Method: Compositional matrix adjust.
Identities = 68/100 (68%), Positives = 81/100 (81%), Gaps = 2/100 (2%)
Query 17 VNIPKTRNTFCRGSKCRKHTPHKVSQYKKGKDSLVVQGKRRYDAKQKGFGGQTKPIFRKK 76
VN+PK R TFC+ KC+ H HKV+QYKK K+ QG+RRYD KQ+GFGGQTKPIFRKK
Sbjct 20 VNVPKQRRTFCK--KCKVHKLHKVTQYKKSKERKGAQGRRRYDRKQQGFGGQTKPIFRKK 77
Query 77 AKTTKKIVLKLECTKCKTKRLLPIKRCKSFELGADKKAKG 116
AKTTKKIVL++ECT+CK ++ P+KRCK FELG DKK KG
Sbjct 78 AKTTKKIVLRMECTECKYRKQTPLKRCKHFELGGDKKRKG 117
> YNL162w
Length=106
Score = 134 bits (336), Expect = 6e-32, Method: Compositional matrix adjust.
Identities = 73/101 (72%), Positives = 83/101 (82%), Gaps = 0/101 (0%)
Query 16 MVNIPKTRNTFCRGSKCRKHTPHKVSQYKKGKDSLVVQGKRRYDAKQKGFGGQTKPIFRK 75
MVN+PKTR T+C+G CRKHT HKV+QYK GK SL QGKRRYD KQ GFGGQTKP+F K
Sbjct 1 MVNVPKTRKTYCKGKTCRKHTQHKVTQYKAGKASLFAQGKRRYDRKQSGFGGQTKPVFHK 60
Query 76 KAKTTKKIVLKLECTKCKTKRLLPIKRCKSFELGADKKAKG 116
KAKTTKK+VL+LEC KCKT+ L +KRCK FELG +KK KG
Sbjct 61 KAKTTKKVVLRLECVKCKTRAQLTLKRCKHFELGGEKKQKG 101
> YHR141c
Length=106
Score = 134 bits (336), Expect = 6e-32, Method: Compositional matrix adjust.
Identities = 73/101 (72%), Positives = 83/101 (82%), Gaps = 0/101 (0%)
Query 16 MVNIPKTRNTFCRGSKCRKHTPHKVSQYKKGKDSLVVQGKRRYDAKQKGFGGQTKPIFRK 75
MVN+PKTR T+C+G CRKHT HKV+QYK GK SL QGKRRYD KQ GFGGQTKP+F K
Sbjct 1 MVNVPKTRKTYCKGKTCRKHTQHKVTQYKAGKASLFAQGKRRYDRKQSGFGGQTKPVFHK 60
Query 76 KAKTTKKIVLKLECTKCKTKRLLPIKRCKSFELGADKKAKG 116
KAKTTKK+VL+LEC KCKT+ L +KRCK FELG +KK KG
Sbjct 61 KAKTTKKVVLRLECVKCKTRAQLTLKRCKHFELGGEKKQKG 101
> SPAC15E1.03
Length=106
Score = 129 bits (324), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 71/102 (69%), Positives = 80/102 (78%), Gaps = 0/102 (0%)
Query 16 MVNIPKTRNTFCRGSKCRKHTPHKVSQYKKGKDSLVVQGKRRYDAKQKGFGGQTKPIFRK 75
MVNIPKTR T+C G CRKHT H+V+QYKKG DS + QGKRRYD KQ GFGGQTKP+F K
Sbjct 1 MVNIPKTRKTYCPGKNCRKHTVHRVTQYKKGPDSKLAQGKRRYDRKQSGFGGQTKPVFHK 60
Query 76 KAKTTKKIVLKLECTKCKTKRLLPIKRCKSFELGADKKAKGG 117
KAK TKK+VL+LEC CK K L +KRCK FELG +KK KG
Sbjct 61 KAKVTKKVVLRLECVSCKYKNQLVLKRCKHFELGGEKKTKGA 102
> ECU10g1300
Length=104
Score = 118 bits (296), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 62/101 (61%), Positives = 74/101 (73%), Gaps = 2/101 (1%)
Query 16 MVNIPKTRNTFCRGSKCRKHTPHKVSQYKKGKDSLVVQGKRRYDAKQKGFGGQTKPIFRK 75
MVNIPKTRNTFC+ KC+ ++ HKVSQ KK KD+ QG RRY KQ+G+GGQTKPI R+
Sbjct 1 MVNIPKTRNTFCK--KCKGYSAHKVSQAKKSKDNPRAQGNRRYARKQRGYGGQTKPILRR 58
Query 76 KAKTTKKIVLKLECTKCKTKRLLPIKRCKSFELGADKKAKG 116
KAK TKK+VL LECTKCK P+KR K G ++K KG
Sbjct 59 KAKVTKKLVLNLECTKCKFSHQKPLKRAKHVIFGGERKRKG 99
> Hs22047108
Length=135
Score = 113 bits (283), Expect = 7e-26, Method: Compositional matrix adjust.
Identities = 60/98 (61%), Positives = 74/98 (75%), Gaps = 4/98 (4%)
Query 16 MVNIPKTRNTFCRGSKCRKHTPHKVSQYKKGKDSLVVQGKRRYDAKQKGFGGQTKPIFRK 75
MVN+PKT TFC +KC KH HKV+Q + GKDSL QGK+ Y+ K G+ GQT+PIFRK
Sbjct 1 MVNVPKTHRTFC--NKCVKHQSHKVTQNEMGKDSLYAQGKKCYNRKPSGYCGQTEPIFRK 58
Query 76 KAKTTKKIVLKLECTK--CKTKRLLPIKRCKSFELGAD 111
KAKTTKKIVL+LEC + C++ ++L IKR K FELG D
Sbjct 59 KAKTTKKIVLRLECIEPNCRSTKVLAIKRYKDFELGGD 96
> Hs22066131
Length=105
Score = 112 bits (279), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 67/103 (65%), Positives = 80/103 (77%), Gaps = 5/103 (4%)
Query 16 MVNIPKTRNTFCRGSKCRKHTPHKVSQYKKGKDSLVVQGKRRYDAKQKGFGGQTKPIFRK 75
MVN+PKTR TFC+ KC KH PHKV+QYK KDSL QGKRRYD KQ G+GGQ +P+F K
Sbjct 1 MVNVPKTRRTFCK--KCGKHQPHKVTQYK-SKDSLYAQGKRRYDRKQSGYGGQRRPVFLK 57
Query 76 KAKTTKKIVLKLECTK--CKTKRLLPIKRCKSFELGADKKAKG 116
KAKTTKK VL+ EC + C++KR++ I+RCK FE G DKK KG
Sbjct 58 KAKTTKKTVLRPECVEPNCRSKRMVVIQRCKHFEPGGDKKRKG 100
> Hs20555200
Length=143
Score = 98.6 bits (244), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 61/104 (58%), Positives = 77/104 (74%), Gaps = 4/104 (3%)
Query 16 MVNIPKTRNTFCRGSKCRKHTPHKVSQYKKGKDSLVVQGKRRYDAKQKGFGGQTKPIFRK 75
+V++PKT+ FC+ KC KH PHKV+QYKKGK+ L QGK+ YD K G+ GQTK IF+K
Sbjct 41 IVSVPKTQWIFCK--KCGKHQPHKVTQYKKGKNCLYAQGKQPYDGKHSGYDGQTKRIFQK 98
Query 76 KAKTTKKIVLKLECTK--CKTKRLLPIKRCKSFELGADKKAKGG 117
KAKTTKK VL+LE + C +KR+L IK C+ FELG DKK +G
Sbjct 99 KAKTTKKTVLRLERVEPNCISKRMLAIKICEHFELGGDKKKRGA 142
> Hs22066175_1
Length=445
Score = 70.9 bits (172), Expect = 6e-13, Method: Composition-based stats.
Identities = 45/100 (45%), Positives = 57/100 (57%), Gaps = 8/100 (8%)
Query 16 MVNIPKTRNTFCRGSKCRKHTPHKVSQYKKGKDSLVVQGKRRYDAKQKGFGGQTKPIFRK 75
M+NI K+ TFCR K KH HKV+Q KKGKDSL QGK+ YD KQ G+ GQTK IF+K
Sbjct 1 MMNIHKSHWTFCR--KYGKHQSHKVTQCKKGKDSLYAQGKQCYDRKQSGYDGQTKLIFQK 58
Query 76 KAKTTKKIVLKLECTKCKTKRLLPIKRCKSFELGADKKAK 115
+ K +C K T+ L +L DK+ +
Sbjct 59 RLNH------KEDCEKVDTRNLENFLENMGIKLTEDKEMQ 92
> Hs22058363
Length=93
Score = 68.2 bits (165), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 45/69 (65%), Positives = 54/69 (78%), Gaps = 2/69 (2%)
Query 49 SLVVQGKRRYDAKQKGFGGQTKPIFRKKAKTTKKIVLKLECTK--CKTKRLLPIKRCKSF 106
S + +GKR YD KQ G+GGQTKPIF KKAKTTKK+VL+LEC + C++KR+L I RCK F
Sbjct 19 SSIQEGKRCYDQKQSGYGGQTKPIFWKKAKTTKKVVLRLECVEPNCRSKRMLAINRCKHF 78
Query 107 ELGADKKAK 115
ELG KK K
Sbjct 79 ELGGYKKRK 87
> Hs17442298
Length=211
Score = 59.7 bits (143), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 33/60 (55%), Positives = 45/60 (75%), Gaps = 3/60 (5%)
Query 64 GFGGQTKPIFRKK-AKTTKKIVLKLECT--KCKTKRLLPIKRCKSFELGADKKAKGGPQF 120
G+GGQTK IF K KTTKKIV++L+C KC++KR+L IK+CK+F+LG D++ QF
Sbjct 152 GYGGQTKQIFWKSLCKTTKKIVMRLKCVEPKCRSKRMLTIKKCKNFKLGRDERKSQMIQF 211
> Hs18573385_2
Length=65
Score = 59.3 bits (142), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 26/39 (66%), Positives = 33/39 (84%), Gaps = 0/39 (0%)
Query 54 GKRRYDAKQKGFGGQTKPIFRKKAKTTKKIVLKLECTKC 92
GK+ YD K G+GGQ+KPIF+KKAKTT+KIVL+LEC +
Sbjct 1 GKQHYDRKHSGYGGQSKPIFQKKAKTTEKIVLRLECVEL 39
> Hs22048689
Length=193
Score = 58.5 bits (140), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 31/50 (62%), Positives = 35/50 (70%), Gaps = 3/50 (6%)
Query 16 MVNIPKTRNTFCRGSKCRKHTPHKVSQYKKGKDSLVVQGKRRYDAKQKGF 65
MVN+PKTR FC+ KC KH P+KV QYK GKDS QGK R D KQ G+
Sbjct 1 MVNVPKTRRAFCK--KCGKHQPYKVPQYK-GKDSSYAQGKMRCDQKQSGY 47
> Hs18601762
Length=136
Score = 55.8 bits (133), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 27/35 (77%), Positives = 30/35 (85%), Gaps = 0/35 (0%)
Query 54 GKRRYDAKQKGFGGQTKPIFRKKAKTTKKIVLKLE 88
GK YD KQ G+GGQTKPIF KKAKTTKKIVL+L+
Sbjct 70 GKWHYDRKQSGYGGQTKPIFWKKAKTTKKIVLRLD 104
> Hs17437316
Length=224
Score = 42.0 bits (97), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 24/73 (32%), Positives = 36/73 (49%), Gaps = 2/73 (2%)
Query 14 LKMVNIPKTRNTFCRGSKCRKHTPHKVSQYKKGKDSLVVQGKRRYDAKQKGFGGQTKPIF 73
L+ V +P R TF + KC H PH+ ++YKKGKDSL Q + + G Q
Sbjct 65 LQDVRVPGNRRTFYK--KCGTHQPHRATRYKKGKDSLSAQRRSAVMGNRVAMGKQQLLFL 122
Query 74 RKKAKTTKKIVLK 86
+ T++ + K
Sbjct 123 LGEISTSEGVAQK 135
> Hs22056645
Length=559
Score = 29.3 bits (64), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 13/29 (44%), Positives = 21/29 (72%), Gaps = 2/29 (6%)
Query 4 ADWKRGKIWILKMVNIPKTRNTFCRGSKC 32
A ++G+IWIL+++ I K+ FCR S+C
Sbjct 224 AQGRKGRIWILEVLAIGKS--VFCRKSRC 250
> At2g30010
Length=398
Score = 28.1 bits (61), Expect = 4.5, Method: Composition-based stats.
Identities = 17/60 (28%), Positives = 28/60 (46%), Gaps = 3/60 (5%)
Query 12 WILKMVNIPKTRNTFCRGSKCRKHTPHKVSQYKKGKDSLVVQGKRRYDAKQKGFGGQTKP 71
W+L+ +N P TR F S + P++ + + K S + QG + + F G T P
Sbjct 260 WVLRYINSPLTRVFFLSVSPTH-YNPNEWTS--RSKTSTITQGGKSCYGQTTPFSGTTYP 316
> Hs4758164
Length=674
Score = 27.3 bits (59), Expect = 6.6, Method: Composition-based stats.
Identities = 20/73 (27%), Positives = 29/73 (39%), Gaps = 10/73 (13%)
Query 21 KTRNTFCRGSKCRKHTPHKVSQYKKGKDSLVVQGKRRYDAKQKGFGGQTKPIFRKKAKTT 80
KT + R S R+ HK S K GKD L + F + P+F
Sbjct 433 KTLSAAARRSFFRRKHKHKRSGSKDGKDLLALD----------AFSSDSIPLFEDSVSLA 482
Query 81 KKIVLKLECTKCK 93
+ V K++CT +
Sbjct 483 YQRVQKVDCTALR 495
> Hs22046315
Length=872
Score = 27.3 bits (59), Expect = 6.6, Method: Composition-based stats.
Identities = 22/83 (26%), Positives = 36/83 (43%), Gaps = 4/83 (4%)
Query 37 PHKVSQYKKGKDSLVVQGKRRYDAKQKGFGGQT-KP---IFRKKAKTTKKIVLKLECTKC 92
P +S ++G +L V KR+ Q + KP +F K TT+ CT+
Sbjct 476 PWGLSLNQRGSKTLAVGLKRQNCKNQDSLTASSCKPNRMLFSDKLVTTEDGQQHPGCTET 535
Query 93 KTKRLLPIKRCKSFELGADKKAK 115
R LP + C LG++ + +
Sbjct 536 HAHRGLPFQSCSRQHLGSESEVR 558
> Hs22052226
Length=674
Score = 27.3 bits (59), Expect = 7.1, Method: Composition-based stats.
Identities = 20/73 (27%), Positives = 29/73 (39%), Gaps = 10/73 (13%)
Query 21 KTRNTFCRGSKCRKHTPHKVSQYKKGKDSLVVQGKRRYDAKQKGFGGQTKPIFRKKAKTT 80
KT + R S R+ HK S K GKD L + F + P+F
Sbjct 433 KTLSAAARRSFFRRKHKHKRSGSKDGKDLLALD----------AFSSDSIPLFEDSVSLA 482
Query 81 KKIVLKLECTKCK 93
+ V K++CT +
Sbjct 483 YQRVQKVDCTALR 495
Lambda K H
0.321 0.136 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1160968786
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40