bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0907_orf1
Length=269
Score E
Sequences producing significant alignments: (Bits) Value
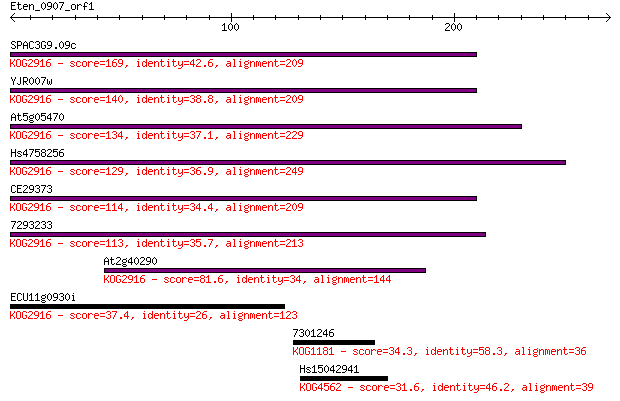
SPAC3G9.09c 169 4e-42
YJR007w 140 2e-33
At5g05470 134 2e-31
Hs4758256 129 9e-30
CE29373 114 1e-25
7293233 113 4e-25
At2g40290 81.6 2e-15
ECU11g0930i 37.4 0.036
7301246 34.3 0.28
Hs15042941 31.6 1.6
> SPAC3G9.09c
Length=306
Score = 169 bits (429), Expect = 4e-42, Method: Compositional matrix adjust.
Identities = 89/210 (42%), Positives = 127/210 (60%), Gaps = 32/210 (15%)
Query 1 IDLSKSRVSPEDIVKCEERYSKGKKVHQTVRHVALKHNLGVDELNAKVIWPLYKKYGHAL 60
IDLSK RVSPED+VKCEER++K K VH +RH+A KHN+ ++ + + WPLY+KYGHA
Sbjct 83 IDLSKRRVSPEDVVKCEERFNKSKAVHSIMRHIAEKHNVPLETMYTTIGWPLYRKYGHAY 142
Query 61 DALKAAALDPEEVFGGLDVSEE-VKKSLIQDIQLRLAPQALKLRARIDVWCFGKDGIDAV 119
DA K A +P+ VF GL+ + V L+ I RL PQ +K+RA ++V CFG +GI+A+
Sbjct 143 DAFKLAISNPDHVFEGLEPPKSGVINDLLAQISRRLTPQPIKIRADVEVTCFGYEGINAI 202
Query 120 KKALLAAKEKAAETPEKAAETPEKAAETPEKAAETPERAVDTPQTQDIAVKLIAPPQYVV 179
K AL AA++ E E P I VKL+APP YV+
Sbjct 203 KAALKAAEDVHTE--------------------EVP-----------IKVKLVAPPLYVL 231
Query 180 VTSSFDKESGMQKIQQAISRISQTIKSYRG 209
+T++ DK G++K+++AI I ++I + G
Sbjct 232 LTNALDKSLGLKKLEEAIGAIEKSITASNG 261
> YJR007w
Length=304
Score = 140 bits (354), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 81/210 (38%), Positives = 122/210 (58%), Gaps = 33/210 (15%)
Query 1 IDLSKSRVSPEDIVKCEERYSKGKKVHQTVRHVALKHNLGVDELNAKVIWPLYKKYGHAL 60
IDLSK RVS EDI+KCEE+Y K K VH +R+ A K + ++EL + WPL +K+GHA
Sbjct 83 IDLSKRRVSSEDIIKCEEKYQKSKTVHSILRYCAEKFQIPLEELYKTIAWPLSRKFGHAY 142
Query 61 DALKAAALDPEEVFGGLDV-SEEVKKSLIQDIQLRLAPQALKLRARIDVWCFGKDGIDAV 119
+A K + +D E V+ G++ S++V L I RL PQA+K+RA ++V CF +GIDA+
Sbjct 143 EAFKLSIID-ETVWEGIEPPSKDVLDELKNYISKRLTPQAVKIRADVEVSCFSYEGIDAI 201
Query 120 KKALLAAKEKAAETPEKAAETPEKAAETPEKAAETPERAVDTPQTQDIAVKLIAPPQYVV 179
K AL +A++ + E Q Q + VKL+A P YV+
Sbjct 202 KDALKSAEDMSTE------------------------------QMQ-VKVKLVAAPLYVL 230
Query 180 VTSSFDKESGMQKIQQAISRISQTIKSYRG 209
T + DK+ G+++++ AI +I++ I Y G
Sbjct 231 TTQALDKQKGIEQLESAIEKITEVITKYGG 260
> At5g05470
Length=344
Score = 134 bits (336), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 85/248 (34%), Positives = 124/248 (50%), Gaps = 52/248 (20%)
Query 1 IDLSKSRVSPEDIVKCEERYSKGKKVHQTVRHVALKHNLGVDELNAKVIWPLYKKYGHAL 60
IDLSK RVS ED CEERY+K K VH +RHVA + ++EL + WPLYKK+GHA
Sbjct 87 IDLSKRRVSDEDKEACEERYNKSKLVHSIMRHVAETVGVDLEELYVNIGWPLYKKHGHAF 146
Query 61 DALKAAALDPEEVFGGLD-------------------VSEEVKKSLIQDIQLRLAPQALK 101
+A K DP+ VF L VSEE+K + ++DI+ R+ PQ +K
Sbjct 147 EAFKIVVTDPDSVFDALTREVKETGPDGVEVTKVVPAVSEELKDAFLKDIRRRMTPQPMK 206
Query 102 LRARIDVWCFGKDGIDAVKKALLAAKEKAAETPEKAAETPEKAAETPEKAAETPERAVDT 161
+RA I++ CF DG+ +K+A+ A+ + D
Sbjct 207 IRADIELKCFQFDGVLHIKEAMKKAEAVGTD---------------------------DC 239
Query 162 PQTQDIAVKLIAPPQYVVVTSSFDKESGMQKIQQAISRISQTIKSYRGGDFKQQGEIMVL 221
P + +KL+APP YV+ T + KE G+ + +AI I+ ++G ++G V
Sbjct 240 P----VKIKLVAPPLYVLTTHTHYKEKGIVTLNKAIEACITAIEEHKGKLVVKEGARAV- 294
Query 222 GSEEDRRL 229
SE D +L
Sbjct 295 -SERDDKL 301
> Hs4758256
Length=315
Score = 129 bits (323), Expect = 9e-30, Method: Compositional matrix adjust.
Identities = 92/261 (35%), Positives = 140/261 (53%), Gaps = 45/261 (17%)
Query 1 IDLSKSRVSPEDIVKCEERYSKGKKVHQTVRHVALKHNLGVDE----LNAKVIWPL---Y 53
IDLSK RVSPE+ +KCE++++K K V+ +RHVA DE L + W Y
Sbjct 83 IDLSKRRVSPEEAIKCEDKFTKSKTVYSILRHVAEVLEYTKDEQLESLFQRTAWVFDDKY 142
Query 54 KKYGH-ALDALKAAALDPEEVFGGLDVSEEVKKSLIQDIQLRLAPQALKLRARIDVWCFG 112
K+ G+ A DA K A DP + LD++E+ ++ LI +I RL PQA+K+RA I+V C+G
Sbjct 143 KRPGYGAYDAFKHAVSDPS-ILDSLDLNEDEREVLINNINRRLTPQAVKIRADIEVACYG 201
Query 113 KDGIDAVKKALLAAKEKAAETPEKAAETPEKAAETPEKAAETPERAVDTPQTQDIAVKLI 172
+GIDAVK+AL A + E + P I + LI
Sbjct 202 YEGIDAVKEALRAGLNCSTE---------------------------NMP----IKINLI 230
Query 173 APPQYVVVTSSFDKESGMQKIQQAISRISQTIKSYRGGDFKQQGEIMVLG----SEEDRR 228
APP+YV+ T++ ++ G+ + QA++ I + I+ RG F Q E V+ +E R+
Sbjct 231 APPRYVMTTTTLERTEGLSVLSQAMAVIKEKIEEKRGV-FNVQMEPKVVTDTDETELARQ 289
Query 229 LEELLLEVSESEEEEEEEEEE 249
+E L E +E + +++ EE E
Sbjct 290 MERLERENAEVDGDDDAEEME 310
> CE29373
Length=342
Score = 114 bits (286), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 72/216 (33%), Positives = 109/216 (50%), Gaps = 39/216 (18%)
Query 1 IDLSKSRVSPEDIVKCEERYSKGKKVHQTVRHVALKHNLGVDE----LNAKVIWPL---Y 53
IDLSK RV +D+ +C+ER++ K V+ +RHVA + DE L K W
Sbjct 80 IDLSKRRVYQKDLKQCDERFANAKMVNSILRHVAEQVGYTTDEELEDLYQKTAWHFDRKE 139
Query 54 KKYGHALDALKAAALDPEEVFGGLDVSEEVKKSLIQDIQLRLAPQALKLRARIDVWCFGK 113
K+ + DA K A +P + D+S ++K+ L++DI+ +L PQA+K+RA I+V CF
Sbjct 140 KRKAASYDAFKKAITEPT-ILDECDISADIKEKLLEDIRKKLTPQAVKIRADIEVSCFDY 198
Query 114 DGIDAVKKALLAAKEKAAETPEKAAETPEKAAETPEKAAETPERAVDTPQTQDIAVKLIA 173
DGIDAVK AL+A K + T P I + LIA
Sbjct 199 DGIDAVKAALIAGKNCSNGT------FP-------------------------IKINLIA 227
Query 174 PPQYVVVTSSFDKESGMQKIQQAISRISQTIKSYRG 209
P +VV T + D+E G++ + + I I+ ++G
Sbjct 228 APHFVVTTQTLDREGGIEAVNSILDTIKNAIEGFKG 263
> 7293233
Length=341
Score = 113 bits (283), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 76/220 (34%), Positives = 110/220 (50%), Gaps = 40/220 (18%)
Query 1 IDLSKSRVSPEDIVKCEERYSKGKKVHQTVRHVA----LKHNLGVDELNAKVIWPLYKKY 56
IDLSK RVSPED+ KC ER++K K ++ +RHVA + N +++L K W KKY
Sbjct 82 IDLSKRRVSPEDVEKCTERFAKAKAINSLLRHVADILGFEGNEKLEDLYQKTAWHFEKKY 141
Query 57 GH---ALDALKAAALDPEEVFGGLDVSEEVKKSLIQDIQLRLAPQALKLRARIDVWCFGK 113
+ A D K + DP VF ++ E K+ L+ +I+ +L +K+RA I+ C+G
Sbjct 142 NNKTVAYDIFKQSVTDP-TVFDECNLEPETKEVLLSNIKRKLVSPTVKIRADIECSCYGY 200
Query 114 DGIDAVKKALLAAKEKAAETPEKAAETPEKAAETPEKAAETPERAVDTPQTQDIAVKLIA 173
+GIDAVK +L E + E E P I + LIA
Sbjct 201 EGIDAVKASLTKGLELSTE------ELP-------------------------IRINLIA 229
Query 174 PPQYVVVTSSFDKESGMQKIQQAISRISQTIKSYRGGDFK 213
PP YV+ TS+ K G++ ++ AI I Y G+FK
Sbjct 230 PPLYVMTTSTTKKTDGLKALEVAIEHIRAKTSEY-DGEFK 268
> At2g40290
Length=154
Score = 81.6 bits (200), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 49/163 (30%), Positives = 77/163 (47%), Gaps = 50/163 (30%)
Query 43 ELNAKVIWPLYKKYGHALDALKAAALDPEEVFGGLD-------------------VSEEV 83
+L + WPLY+++GHA +A K DP+ V G L V+EEV
Sbjct 4 DLYVNIGWPLYRRHGHAFEAFKILVTDPDSVLGPLTREIKEVGPDGQEVTKVVPAVTEEV 63
Query 84 KKSLIQDIQLRLAPQALKLRARIDVWCFGKDGIDAVKKALLAAKEKAAETPEKAAETPEK 143
K +L+++I+ R+ PQ +K+RA I++ CF DG+ +K+A+ A+ E
Sbjct 64 KDALVKNIRRRMTPQPMKIRADIELKCFQFDGVVHIKEAMKNAEAAGNE----------- 112
Query 144 AAETPEKAAETPERAVDTPQTQDIAVKLIAPPQYVVVTSSFDK 186
D P + +KL+APP YV+ T + DK
Sbjct 113 ----------------DCP----VKIKLVAPPLYVLTTQTLDK 135
> ECU11g0930i
Length=283
Score = 37.4 bits (85), Expect = 0.036, Method: Compositional matrix adjust.
Identities = 32/123 (26%), Positives = 58/123 (47%), Gaps = 5/123 (4%)
Query 1 IDLSKSRVSPEDIVKCEERYSKGKKVHQTVRHVALKHNLGVDELNAKVIWPLYKKYGHAL 60
IDLS S+V+ + +C E +K K + + A + + V EL ++ + +++G +L
Sbjct 83 IDLSMSKVTENEKSECRETSAKNKLAYHIMVKAAKRLGMEVSELYERMGYDKEEEFG-SL 141
Query 61 DALKAAALDPEEVFGGLDVSEEVKKSLIQDIQLRLAPQALKLRARIDVWCFGKDGIDAVK 120
A D ++ + +KK + + Q K+RA IDV C + GI ++K
Sbjct 142 YYFFARVKDNGDIMDDDAIGVTIKKLIREQFQ----ASTYKVRADIDVVCSSRGGIVSIK 197
Query 121 KAL 123
K
Sbjct 198 KVF 200
> 7301246
Length=2768
Score = 34.3 bits (77), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 21/36 (58%), Positives = 24/36 (66%), Gaps = 0/36 (0%)
Query 128 EKAAETPEKAAETPEKAAETPEKAAETPERAVDTPQ 163
EKA E PE ETPEKA E PE ETPE+A + P+
Sbjct 2388 EKATEQPELEKETPEKATEQPELEKETPEKATEQPE 2423
> Hs15042941
Length=219
Score = 31.6 bits (70), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 18/39 (46%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 131 AETPEKAAETPEKAAETPEKAAETPERAVDTPQTQDIAV 169
A TPE PE+ TPE+A+ TPE A T Q Q +V
Sbjct 41 ASTPEDDLSGPEEDPSTPEEASTTPEEASSTAQAQKPSV 79
Lambda K H
0.308 0.128 0.345
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5737723452
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40