bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0926_orf1
Length=92
Score E
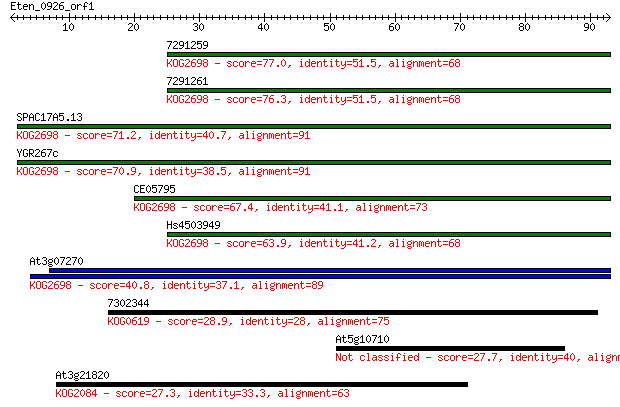
Sequences producing significant alignments: (Bits) Value
7291259 77.0 9e-15
7291261 76.3 1e-14
SPAC17A5.13 71.2 4e-13
YGR267c 70.9 5e-13
CE05795 67.4 6e-12
Hs4503949 63.9 7e-11
At3g07270 40.8 6e-04
7302344 28.9 2.4
At5g10710 27.7 5.4
At3g21820 27.3 7.5
> 7291259
Length=273
Score = 77.0 bits (188), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 35/68 (51%), Positives = 48/68 (70%), Gaps = 2/68 (2%)
Query 25 LLNTPRRAAEAFLYFTKGYELSLEAAVGSGVFAYTEETGDTIVIKDLFVHSVCEHHLLPF 84
L+ TP RAA+A LYFTKGY+ SLE + VF E+ + +V+KD+ + S+CEHHL+PF
Sbjct 113 LIKTPERAAKAMLYFTKGYDQSLEDVLNGAVF--DEDHDEMVVVKDIEMFSMCEHHLVPF 170
Query 85 HGTCSIAY 92
+G SI Y
Sbjct 171 YGKVSIGY 178
> 7291261
Length=308
Score = 76.3 bits (186), Expect = 1e-14, Method: Composition-based stats.
Identities = 35/68 (51%), Positives = 48/68 (70%), Gaps = 2/68 (2%)
Query 25 LLNTPRRAAEAFLYFTKGYELSLEAAVGSGVFAYTEETGDTIVIKDLFVHSVCEHHLLPF 84
L+ TP RAA+A LYFTKGY+ SLE + VF E+ + +V+KD+ + S+CEHHL+PF
Sbjct 148 LIKTPERAAKAMLYFTKGYDQSLEDVLNGAVF--DEDHDEMVVVKDIEMFSMCEHHLVPF 205
Query 85 HGTCSIAY 92
+G SI Y
Sbjct 206 YGKVSIGY 213
> SPAC17A5.13
Length=235
Score = 71.2 bits (173), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 37/91 (40%), Positives = 55/91 (60%), Gaps = 3/91 (3%)
Query 2 KRMEQAVSDLLKALPAGVLSEEVLLNTPRRAAEAFLYFTKGYELSLEAAVGSGVFAYTEE 61
K++ A+S +L+ L + LL TP R A+A LYFTKGYE +L + VF E+
Sbjct 53 KKISNAISTILECLGEDP-ERQGLLGTPERYAKAMLYFTKGYEQNLTEVINEAVFQ--ED 109
Query 62 TGDTIVIKDLFVHSVCEHHLLPFHGTCSIAY 92
+ ++++D+ V S+CEHHL+PF G I Y
Sbjct 110 HEEMVIVRDIDVFSLCEHHLVPFIGKIHIGY 140
> YGR267c
Length=243
Score = 70.9 bits (172), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 35/92 (38%), Positives = 60/92 (65%), Gaps = 4/92 (4%)
Query 2 KRMEQAVSDLLKALPAGVLSEEVLLNTPRRAAEAFLYFTKGYELS-LEAAVGSGVFAYTE 60
+R+ A+ +L L V + E LL+TP+R A+A LYFTKGY+ + ++ + + VF E
Sbjct 59 QRISGAIKTILTELGEDV-NREGLLDTPQRYAKAMLYFTKGYQTNIMDDVIKNAVFE--E 115
Query 61 ETGDTIVIKDLFVHSVCEHHLLPFHGTCSIAY 92
+ + ++++D+ ++S+CEHHL+PF G I Y
Sbjct 116 DHDEMVIVRDIEIYSLCEHHLVPFFGKVHIGY 147
> CE05795
Length=223
Score = 67.4 bits (163), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 30/73 (41%), Positives = 47/73 (64%), Gaps = 2/73 (2%)
Query 20 LSEEVLLNTPRRAAEAFLYFTKGYELSLEAAVGSGVFAYTEETGDTIVIKDLFVHSVCEH 79
++ + LL TP RAA+A + FTKGY+ L+ + VF E+ + +++KD+ + S+CEH
Sbjct 58 INRQGLLKTPERAAKAMMAFTKGYDDQLDELLNEAVF--DEDHDEMVIVKDIEMFSLCEH 115
Query 80 HLLPFHGTCSIAY 92
HL+PF G I Y
Sbjct 116 HLVPFMGKVHIGY 128
> Hs4503949
Length=250
Score = 63.9 bits (154), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 28/68 (41%), Positives = 43/68 (63%), Gaps = 2/68 (2%)
Query 25 LLNTPRRAAEAFLYFTKGYELSLEAAVGSGVFAYTEETGDTIVIKDLFVHSVCEHHLLPF 84
LL TP RAA A +FTKGY+ ++ + +F E+ + +++KD+ + S+CEHHL+PF
Sbjct 91 LLKTPWRAASAMQFFTKGYQETISDVLNDAIF--DEDHDEMVIVKDIDMFSMCEHHLVPF 148
Query 85 HGTCSIAY 92
G I Y
Sbjct 149 VGKVHIGY 156
> At3g07270
Length=466
Score = 40.8 bits (94), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 32/98 (32%), Positives = 48/98 (48%), Gaps = 12/98 (12%)
Query 7 AVSDLLKALPAGV---LSEEVLLNTPRRAAEAFLYFTKGYELSLEAAVGSGVFA------ 57
A+ D +K L G+ ++ E + TP R A+A T+GY+ ++ V S +F
Sbjct 34 AIQDAVKLLLQGLHEDVNREGIKKTPFRVAKALREGTRGYKQKVKDYVQSALFPEAGLDE 93
Query 58 ---YTEETGDTIVIKDLFVHSVCEHHLLPFHGTCSIAY 92
G +V++DL +S CE LLPFH C I Y
Sbjct 94 GVGQAGGVGGLVVVRDLDHYSYCESCLLPFHVKCHIGY 131
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 31/90 (34%), Positives = 44/90 (48%), Gaps = 3/90 (3%)
Query 4 MEQAVSDLLKALPAGVLSEEVLLNTPRRAAEAFLYFTK-GYELSLEAAVGSGVFAYTEET 62
M AV +LK+L L +E L+ TP R + L F + E+ L + + V +E
Sbjct 270 MVSAVVSILKSLGEDPLRKE-LIATPTRFLKWMLNFQRTNLEMKLNSFNPAKVNGEVKEK 328
Query 63 GDTIVIKDLFVHSVCEHHLLPFHGTCSIAY 92
+ F S+CEHHLLPF+G I Y
Sbjct 329 RLHCELNMPF-WSMCEHHLLPFYGVVHIGY 357
> 7302344
Length=799
Score = 28.9 bits (63), Expect = 2.4, Method: Composition-based stats.
Identities = 21/75 (28%), Positives = 36/75 (48%), Gaps = 2/75 (2%)
Query 16 PAGVLSEEVLLNTPRRAAEAFLYFTKGYELSLEAAVGSGVFAYTEETGDTIVIKDLFVHS 75
P LSEE++L+ + + L T LS + + +F Y EE + I+I + +H
Sbjct 521 PIKDLSEEIILHELHFSTDYGLILTWLLNLSKKDYMCDAIFVYKEEHINEILIDNSPIH- 579
Query 76 VCEHHLLPFHGTCSI 90
CE ++ T S+
Sbjct 580 -CESKVVNGQNTVSV 593
> At5g10710
Length=230
Score = 27.7 bits (60), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 14/36 (38%), Positives = 21/36 (58%), Gaps = 1/36 (2%)
Query 51 VGSGVFAYTEETGDTIVIKDLFVHSVCE-HHLLPFH 85
VG + AY + +IK+LF H + E +H LP+H
Sbjct 102 VGDLLQAYVDRKEQVRLIKELFGHQISEIYHSLPYH 137
> At3g21820
Length=460
Score = 27.3 bits (59), Expect = 7.5, Method: Composition-based stats.
Identities = 21/64 (32%), Positives = 29/64 (45%), Gaps = 9/64 (14%)
Query 8 VSDLLKALPAGVLSEEV-LLNTPRRAAEAFLYFTKGYELSLEAAVGSGVFAYTEETGDTI 66
V+ LL LP L E L T RR G E+ +G GV+A +E D +
Sbjct 15 VAALLAPLPTPQLQEYFNKLITSRRC--------NGIEVKNNGTIGKGVYANSEFDEDEL 66
Query 67 VIKD 70
++KD
Sbjct 67 ILKD 70
Lambda K H
0.321 0.135 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1171209254
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40