bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0935_orf1
Length=173
Score E
Sequences producing significant alignments: (Bits) Value
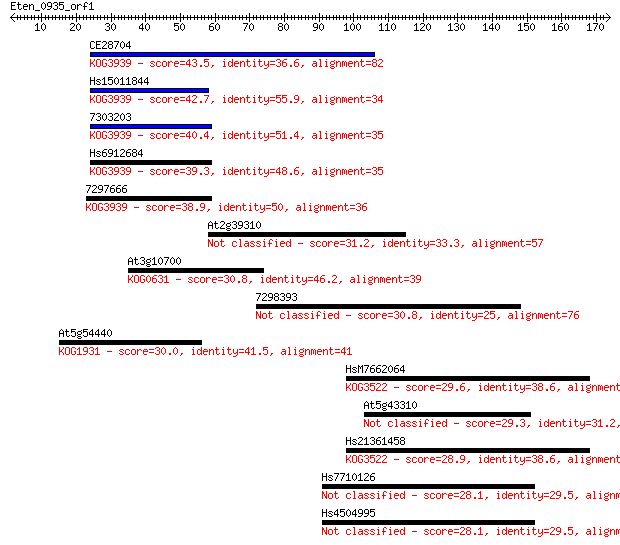
CE28704 43.5 2e-04
Hs15011844 42.7 4e-04
7303203 40.4 0.002
Hs6912684 39.3 0.004
7297666 38.9 0.005
At2g39310 31.2 1.1
At3g10700 30.8 1.2
7298393 30.8 1.4
At5g54440 30.0 2.5
HsM7662064 29.6 2.9
At5g43310 29.3 3.9
Hs21361458 28.9 4.7
Hs7710126 28.1 8.1
Hs4504995 28.1 8.9
> CE28704
Length=378
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 30/89 (33%), Positives = 42/89 (47%), Gaps = 12/89 (13%)
Query 24 LAQSVDFFRLFTGDLYTMGRVAAAHALSDLYACGAEP------LTALAVTLMQKAPPELQ 77
L Q+ DFF D Y MGRV A+ LSDLYA G L A+A+ L +K Q
Sbjct 78 LVQTTDFFYPLIDDPYIMGRVTCANVLSDLYAMGVSECDNMLMLLAVAIDLNEK-----Q 132
Query 78 ANNLL-QFLCGASSVFSSEGCELSGGHSA 105
+ ++ F+ G G ++ GG +
Sbjct 133 RDIVVPLFIQGFKDAADEAGTKIRGGQTV 161
> Hs15011844
Length=448
Score = 42.7 bits (99), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 19/34 (55%), Positives = 22/34 (64%), Gaps = 0/34 (0%)
Query 24 LAQSVDFFRLFTGDLYTMGRVAAAHALSDLYACG 57
L Q+ DFF D Y MGR+A A+ LSDLYA G
Sbjct 133 LVQTTDFFYPLVEDPYMMGRIACANVLSDLYAMG 166
> 7303203
Length=398
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 18/35 (51%), Positives = 22/35 (62%), Gaps = 0/35 (0%)
Query 24 LAQSVDFFRLFTGDLYTMGRVAAAHALSDLYACGA 58
L Q+ DFF D Y MG++A A+ LSDLYA G
Sbjct 104 LVQTTDFFYPIVDDPYMMGKIACANVLSDLYAMGV 138
> Hs6912684
Length=383
Score = 39.3 bits (90), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 17/35 (48%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 24 LAQSVDFFRLFTGDLYTMGRVAAAHALSDLYACGA 58
L Q+ D+ D Y MGR+A A+ LSDLYA G
Sbjct 82 LVQTTDYIYPIVDDPYMMGRIACANVLSDLYAMGV 116
> 7297666
Length=315
Score = 38.9 bits (89), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 18/36 (50%), Positives = 23/36 (63%), Gaps = 0/36 (0%)
Query 23 LLAQSVDFFRLFTGDLYTMGRVAAAHALSDLYACGA 58
LL Q+VDFF D +GR+A A+ LSD+YA G
Sbjct 15 LLIQTVDFFYPMVNDPELLGRIALANVLSDVYAVGV 50
> At2g39310
Length=458
Score = 31.2 bits (69), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 24/57 (42%), Gaps = 0/57 (0%)
Query 58 AEPLTALAVTLMQKAPPELQANNLLQFLCGASSVFSSEGCELSGGHSAAGDGPSAAG 114
E +T+L + L NLL G VF EG ++ G H AGD A G
Sbjct 92 TEVITSLVFKTSKGRKSPLFGPNLLGITTGTKFVFEDEGKKIVGFHGRAGDAVDALG 148
> At3g10700
Length=412
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 18/41 (43%), Positives = 25/41 (60%), Gaps = 3/41 (7%)
Query 35 TGDLYTMGRVAAAHALSDL--YACGAEPLTALAVTLMQKAP 73
+G+L G++ +A LS + Y CGAEPL L L+ KAP
Sbjct 307 SGNLEEFGKLISASGLSSIENYECGAEPLIQLYKILL-KAP 346
> 7298393
Length=536
Score = 30.8 bits (68), Expect = 1.4, Method: Composition-based stats.
Identities = 19/82 (23%), Positives = 34/82 (41%), Gaps = 6/82 (7%)
Query 72 APPELQANNLLQFLCGASSVFSSEGCELSGGHSAAGDG------PSAAGFCVTGRIFYPS 125
+PP + + +L ++ ++ ++ G AA +G P + FY
Sbjct 435 SPPIFKHDIVLSWIVPKNNKLGKRHAPITFGEKAAANGKDKDQEPEKKSRPSNDQKFYSP 494
Query 126 PPGASSAKTNSSKGGNDEKRRQ 147
P G S K N S G N+ R++
Sbjct 495 PSGKYSNKVNQSYGNNNRTRQR 516
> At5g54440
Length=1280
Score = 30.0 bits (66), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 17/41 (41%), Positives = 22/41 (53%), Gaps = 6/41 (14%)
Query 15 GVTAPPVSLLAQSVDFFRLFTGDLYTMGRVAAAHALSDLYA 55
GV AP + +FFRL GDLY++ RV + L LY
Sbjct 364 GVVAPDIE-----KEFFRL-QGDLYSLSRVKCLYPLKCLYV 398
> HsM7662064
Length=1510
Score = 29.6 bits (65), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 27/77 (35%), Positives = 34/77 (44%), Gaps = 9/77 (11%)
Query 98 ELSGGHSAAGDG-------PSAAGFCVTGRIFYPSPPGASSAKTNSSKGGNDEKRRQQQQ 150
ELSG S+ D P A FC G P +SSA+TN G ++
Sbjct 82 ELSGPESSLTDEGIGADPEPPVAAFCGLGTTGMWRPLSSSSAQTNHHGPGTEDS--LGGW 139
Query 151 ALVAPGKPLAPGFLLRK 167
ALV+P P PG L R+
Sbjct 140 ALVSPETPPTPGALRRR 156
> At5g43310
Length=1221
Score = 29.3 bits (64), Expect = 3.9, Method: Composition-based stats.
Identities = 15/53 (28%), Positives = 26/53 (49%), Gaps = 5/53 (9%)
Query 103 HSAAGDGPSAAGFCVTGRIFYPSPPGASS-----AKTNSSKGGNDEKRRQQQQ 150
HS G P G+ + G +YP PGAS T+ S+ G+ +++ ++
Sbjct 368 HSPPGTFPVFQGYTMQGMPYYPGYPGASPYPSPYPSTDDSRRGSGQRKARKHH 420
> Hs21361458
Length=2063
Score = 28.9 bits (63), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 27/77 (35%), Positives = 34/77 (44%), Gaps = 9/77 (11%)
Query 98 ELSGGHSAAGDG-------PSAAGFCVTGRIFYPSPPGASSAKTNSSKGGNDEKRRQQQQ 150
ELSG S+ D P A FC G P +SSA+TN G ++
Sbjct 635 ELSGPESSLTDEGIGADPEPPVAAFCGLGTTGMWRPLSSSSAQTNHHGPGTEDS--LGGW 692
Query 151 ALVAPGKPLAPGFLLRK 167
ALV+P P PG L R+
Sbjct 693 ALVSPETPPTPGALRRR 709
> Hs7710126
Length=922
Score = 28.1 bits (61), Expect = 8.1, Method: Composition-based stats.
Identities = 18/61 (29%), Positives = 23/61 (37%), Gaps = 4/61 (6%)
Query 91 VFSSEGCELSGGHSAAGDGPSAAGFCVTGRIFYPSPPGASSAKTNSSKGGNDEKRRQQQQ 150
V EG +GG S GPS + PS P AS+ K ++ K Q
Sbjct 754 VAGDEGSSTTGGSSEENKGPSGSAVSRKA----PSKPSASTKKAEGKLSNSNSKDGNMQT 809
Query 151 A 151
A
Sbjct 810 A 810
> Hs4504995
Length=862
Score = 28.1 bits (61), Expect = 8.9, Method: Composition-based stats.
Identities = 18/61 (29%), Positives = 23/61 (37%), Gaps = 4/61 (6%)
Query 91 VFSSEGCELSGGHSAAGDGPSAAGFCVTGRIFYPSPPGASSAKTNSSKGGNDEKRRQQQQ 150
V EG +GG S GPS + PS P AS+ K ++ K Q
Sbjct 754 VAGDEGSSTTGGSSEENKGPSGSAVSRKA----PSKPSASTKKAEGKLSNSNSKDGNMQT 809
Query 151 A 151
A
Sbjct 810 A 810
Lambda K H
0.316 0.133 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2598880752
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40