bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0942_orf3
Length=118
Score E
Sequences producing significant alignments: (Bits) Value
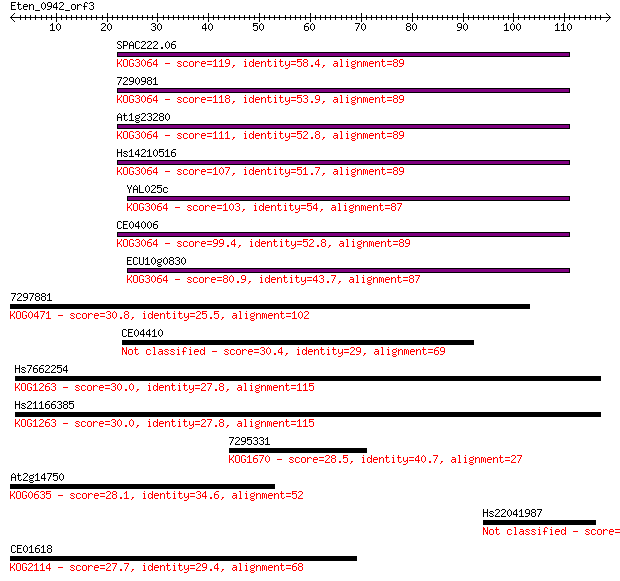
SPAC222.06 119 9e-28
7290981 118 2e-27
At1g23280 111 3e-25
Hs14210516 107 7e-24
YAL025c 103 9e-23
CE04006 99.4 2e-21
ECU10g0830 80.9 5e-16
7297881 30.8 0.56
CE04410 30.4 0.86
Hs7662254 30.0 1.2
Hs21166385 30.0 1.2
7295331 28.5 3.5
At2g14750 28.1 3.6
Hs22041987 27.7 5.2
CE01618 27.7 5.9
> SPAC222.06
Length=302
Score = 119 bits (299), Expect = 9e-28, Method: Compositional matrix adjust.
Identities = 52/89 (58%), Positives = 68/89 (76%), Gaps = 0/89 (0%)
Query 22 MQNDELIWCVVNKHFCAFKKKTDKEDMCRNVYNVTGKCNRVSCPLANSHYATVLEQQGKL 81
MQ DE+IW VV FC+++ K + ++ CRN YNVTG CNR SCPLANS YATV E GKL
Sbjct 1 MQQDEVIWQVVGHEFCSYRIKGEAQNFCRNEYNVTGLCNRQSCPLANSRYATVREDNGKL 60
Query 82 YLCLKTVERAHLPSRLWEKLKLPQSLREA 110
YL +KT+ERAH PS+LW+++KL ++ +A
Sbjct 61 YLYMKTIERAHFPSKLWQRIKLSKNYAKA 89
> 7290981
Length=343
Score = 118 bits (296), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 48/89 (53%), Positives = 69/89 (77%), Gaps = 0/89 (0%)
Query 22 MQNDELIWCVVNKHFCAFKKKTDKEDMCRNVYNVTGKCNRVSCPLANSHYATVLEQQGKL 81
MQ+D+++W ++NK FC+ K KTD CR+ YN+TG C R +CPLANS YATV E++G +
Sbjct 1 MQHDDVVWSIINKSFCSHKVKTDTRTFCRHEYNLTGLCTRRTCPLANSQYATVREEKGII 60
Query 82 YLCLKTVERAHLPSRLWEKLKLPQSLREA 110
YL +KT ERAH+PS+LWE++KL ++ +A
Sbjct 61 YLFIKTAERAHMPSKLWERIKLSRNFEKA 89
> At1g23280
Length=303
Score = 111 bits (278), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 47/89 (52%), Positives = 64/89 (71%), Gaps = 0/89 (0%)
Query 22 MQNDELIWCVVNKHFCAFKKKTDKEDMCRNVYNVTGKCNRVSCPLANSHYATVLEQQGKL 81
MQ+DE+IW V+ C++ K + CRN YNVTG CNR SCPLANS YAT+ + G
Sbjct 1 MQHDEVIWQVIRHKHCSYMAKIETGIFCRNQYNVTGICNRSSCPLANSRYATIRDHDGVF 60
Query 82 YLCLKTVERAHLPSRLWEKLKLPQSLREA 110
YL +KT+ERAH+P++LWE++KLP + +A
Sbjct 61 YLYMKTIERAHMPNKLWERVKLPVNYEKA 89
> Hs14210516
Length=300
Score = 107 bits (266), Expect = 7e-24, Method: Compositional matrix adjust.
Identities = 46/90 (51%), Positives = 67/90 (74%), Gaps = 1/90 (1%)
Query 22 MQNDELIWCVV-NKHFCAFKKKTDKEDMCRNVYNVTGKCNRVSCPLANSHYATVLEQQGK 80
MQ+D++IW + NK FC+FK +T + CRN Y++TG CNR SCPLANS YAT+ E++G+
Sbjct 1 MQSDDVIWDTLGNKQFCSFKIRTKTQSFCRNEYSLTGLCNRSSCPLANSQYATIKEEKGQ 60
Query 81 LYLCLKTVERAHLPSRLWEKLKLPQSLREA 110
YL +K +ERA P RLWE+++L ++ +A
Sbjct 61 CYLYMKVIERAAFPRRLWERVRLSKNYEKA 90
> YAL025c
Length=306
Score = 103 bits (256), Expect = 9e-23, Method: Compositional matrix adjust.
Identities = 47/88 (53%), Positives = 65/88 (73%), Gaps = 1/88 (1%)
Query 24 NDELIWCVVNKHFCAFK-KKTDKEDMCRNVYNVTGKCNRVSCPLANSHYATVLEQQGKLY 82
+DE++W V+N+ FC+ + K + ++ CRN YNVTG C R SCPLANS YATV GKLY
Sbjct 2 SDEIVWQVINQSFCSHRIKAPNGQNFCRNEYNVTGLCTRQSCPLANSKYATVKCDNGKLY 61
Query 83 LCLKTVERAHLPSRLWEKLKLPQSLREA 110
L +KT ERAH P++LWE++KL ++ +A
Sbjct 62 LYMKTPERAHTPAKLWERIKLSKNYTKA 89
> CE04006
Length=323
Score = 99.4 bits (246), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 47/89 (52%), Positives = 60/89 (67%), Gaps = 0/89 (0%)
Query 22 MQNDELIWCVVNKHFCAFKKKTDKEDMCRNVYNVTGKCNRVSCPLANSHYATVLEQQGKL 81
MQ D++ W ++NK CA+K T + C+N N+TG CNR SCPLANS YATV E+ G
Sbjct 1 MQCDDVTWNILNKGQCAYKAWTKPKMFCKNEMNLTGLCNRASCPLANSQYATVREENGVC 60
Query 82 YLCLKTVERAHLPSRLWEKLKLPQSLREA 110
YL K VER+H P RLWEK KL + + +A
Sbjct 61 YLYAKVVERSHYPRRLWEKTKLSKDMNKA 89
> ECU10g0830
Length=233
Score = 80.9 bits (198), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 38/88 (43%), Positives = 58/88 (65%), Gaps = 1/88 (1%)
Query 24 NDELIW-CVVNKHFCAFKKKTDKEDMCRNVYNVTGKCNRVSCPLANSHYATVLEQQGKLY 82
+DE +W + ++ C+FK +T++ +CRN NVTG C+R SCPLANS YATV +L+
Sbjct 2 SDESLWRNISGENHCSFKMRTEENTLCRNQNNVTGLCDRFSCPLANSRYATVRAVGEELF 61
Query 83 LCLKTVERAHLPSRLWEKLKLPQSLREA 110
L +K ER H+P +E+++L + EA
Sbjct 62 LFVKEPERVHVPRDAYEQIRLSSNYEEA 89
> 7297881
Length=584
Score = 30.8 bits (68), Expect = 0.56, Method: Composition-based stats.
Identities = 26/108 (24%), Positives = 49/108 (45%), Gaps = 16/108 (14%)
Query 1 DLKGQAKARK------LDLRRRGELAKMQNDELIWCVVNKHFCAFKKKTDKEDMCRNVYN 54
+L+ Q +AR+ L + +L ++N + VVN+ AFK++ E + N
Sbjct 467 NLRNQQQARRSHYKIYQSLLKLRQLPVLKNGSFVPEVVNRRVFAFKRELKNEHTLLTIVN 526
Query 55 VTGKCNRVSCPLANSHYATVLEQQGKLYLCLKTVERAHLPSRLWEKLK 102
V+ + V A +EQ +L + + V+ H R+ ++LK
Sbjct 527 VSNRTELVDI-------ADFIEQPNRLSVLVAGVDSQH---RVGDRLK 564
> CE04410
Length=467
Score = 30.4 bits (67), Expect = 0.86, Method: Composition-based stats.
Identities = 20/76 (26%), Positives = 31/76 (40%), Gaps = 7/76 (9%)
Query 23 QNDELI-WCVVN------KHFCAFKKKTDKEDMCRNVYNVTGKCNRVSCPLANSHYATVL 75
QND +I WC +N C + E + + YN G R P +H
Sbjct 41 QNDYIINWCNMNGDQLIGARGCVNCNQCGYESVETHSYNRPGAIKRTLVPFVAAHTTINF 100
Query 76 EQQGKLYLCLKTVERA 91
E + ++Y+C ER+
Sbjct 101 ECEIEMYICSGCAERS 116
> Hs7662254
Length=891
Score = 30.0 bits (66), Expect = 1.2, Method: Composition-based stats.
Identities = 32/122 (26%), Positives = 51/122 (41%), Gaps = 25/122 (20%)
Query 2 LKGQAKARKLDLRRRGELAKMQNDEL----IWCVVNKHFCAFKKKTDKEDMCRNVYNVTG 57
L+G K + +A MQ D L I+C H +E R +YNV+
Sbjct 400 LQGMRKGAAMLFPHTFVMAIMQPDNLGTFEIYCQAGSH---------REAGMRAIYNVS- 449
Query 58 KCNRVSCPLANSHYATVLE--QQGKLYLCL-KTVERAHLPSRLWEKLKLPQSLREAKRFV 114
CP H AT + Q ++Y + + VE + P R WE+ QS +++ ++
Sbjct 450 -----QCP---GHQATPRQRYQAARIYYIMAEEVEWDYCPDRSWEREWHNQSEKDSYGYI 501
Query 115 QL 116
L
Sbjct 502 FL 503
> Hs21166385
Length=1158
Score = 30.0 bits (66), Expect = 1.2, Method: Composition-based stats.
Identities = 32/122 (26%), Positives = 51/122 (41%), Gaps = 25/122 (20%)
Query 2 LKGQAKARKLDLRRRGELAKMQNDEL----IWCVVNKHFCAFKKKTDKEDMCRNVYNVTG 57
L+G K + +A MQ D L I+C H +E R +YNV+
Sbjct 667 LQGMRKGAAMLFPHTFVMAIMQPDNLGTFEIYCQAGSH---------REAGMRAIYNVS- 716
Query 58 KCNRVSCPLANSHYATVLE--QQGKLYLCL-KTVERAHLPSRLWEKLKLPQSLREAKRFV 114
CP H AT + Q ++Y + + VE + P R WE+ QS +++ ++
Sbjct 717 -----QCP---GHQATPRQRYQAARIYYIMAEEVEWDYCPDRSWEREWHNQSEKDSYGYI 768
Query 115 QL 116
L
Sbjct 769 FL 770
> 7295331
Length=229
Score = 28.5 bits (62), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 11/27 (40%), Positives = 17/27 (62%), Gaps = 0/27 (0%)
Query 44 DKEDMCRNVYNVTGKCNRVSCPLANSH 70
+ E++C V N+ GK N++S AN H
Sbjct 158 NTEEVCGAVINLRGKSNKISIWTANGH 184
> At2g14750
Length=276
Score = 28.1 bits (61), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 18/52 (34%), Positives = 30/52 (57%), Gaps = 4/52 (7%)
Query 1 DLKGQAKARKLDLRRRGELAKMQNDELIWCVVNKHFCAFKKKTDKEDMCRNV 52
DL +A+ R ++RR GE+AK+ D I C+ + +TD+ D CR++
Sbjct 147 DLSFKAEDRAENIRRVGEVAKLFADAGIICIAS---LISPYRTDR-DACRSL 194
> Hs22041987
Length=376
Score = 27.7 bits (60), Expect = 5.2, Method: Composition-based stats.
Identities = 11/22 (50%), Positives = 14/22 (63%), Gaps = 0/22 (0%)
Query 94 PSRLWEKLKLPQSLREAKRFVQ 115
P LWE+ KL Q+LRE + Q
Sbjct 14 PQNLWEQTKLSQNLREQTKLSQ 35
> CE01618
Length=950
Score = 27.7 bits (60), Expect = 5.9, Method: Composition-based stats.
Identities = 20/69 (28%), Positives = 31/69 (44%), Gaps = 1/69 (1%)
Query 1 DLKGQAKARKLDLRRRGELAKMQNDELIWCVVNKHFCAFKKKTDKEDMCR-NVYNVTGKC 59
D+ G R L L R E ++ ++ KH +K+ D E M +VY++ G+
Sbjct 257 DMDGGEVGRCLQLGRGHEKLQLVASGQYLALLTKHHSLIQKERDSEFMTMLSVYDIKGQY 316
Query 60 NRVSCPLAN 68
SC L N
Sbjct 317 VGFSCSLPN 325
Lambda K H
0.323 0.135 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1167969826
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40