bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0945_orf1
Length=164
Score E
Sequences producing significant alignments: (Bits) Value
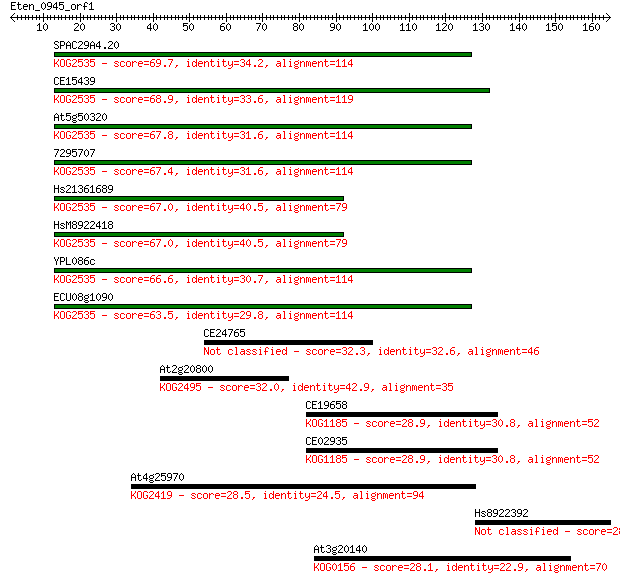
SPAC29A4.20 69.7 2e-12
CE15439 68.9 4e-12
At5g50320 67.8 9e-12
7295707 67.4 1e-11
Hs21361689 67.0 1e-11
HsM8922418 67.0 1e-11
YPL086c 66.6 2e-11
ECU08g1090 63.5 2e-10
CE24765 32.3 0.40
At2g20800 32.0 0.58
CE19658 28.9 4.7
CE02935 28.9 4.9
At4g25970 28.5 5.4
Hs8922392 28.1 7.8
At3g20140 28.1 8.7
> SPAC29A4.20
Length=544
Score = 69.7 bits (169), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 39/119 (32%), Positives = 63/119 (52%), Gaps = 12/119 (10%)
Query 13 CHYCPNEPGQ-----PRSYLSTEPAVLRANQNGWSPLKQFKDRAETLRRNGHVVDKIEVL 67
C YCP P +SY EP +RA + + P +Q + R E LR GH VDK+E +
Sbjct 106 CVYCPGGPDSDFEYSTQSYTGYEPTSMRAIRARYDPYEQARGRVEQLRSLGHTVDKVEYI 165
Query 68 VLGGTWSGYPQDYQEEFIRDVYYAANVFGAPEPVRQPLSLEEEQSLNETADCRIVGLTL 126
++GGT+ P+ Y+ FI +++ A + GA L+E +E ++ + VG+T+
Sbjct 166 IMGGTFMSLPESYRHTFIANLHNALS--GATTE-----DLDEAVKFSEQSETKCVGITI 217
> CE15439
Length=554
Score = 68.9 bits (167), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 40/124 (32%), Positives = 67/124 (54%), Gaps = 13/124 (10%)
Query 13 CHYCPNEPGQ-----PRSYLSTEPAVLRANQNGWSPLKQFKDRAETLRRNGHVVDKIEVL 67
C YCP P +SY EP +RA + ++P Q + R L + GH VDK+E +
Sbjct 112 CVYCPGGPDSDFEYSTQSYTGYEPTSMRAIRARYNPYLQTRGRLNQLMQLGHSVDKVEFI 171
Query 68 VLGGTWSGYPQDYQEEFIRDVYYAANVFGAPEPVRQPLSLEEEQSLNETADCRIVGLTLV 127
V+GGT+ P+DY++ FIR+++ A + + S+EE + +E + + +G+T +
Sbjct 172 VMGGTFMSLPEDYRDFFIRNLHDALSGHTSA-------SVEEAVAYSERSKMKCIGIT-I 223
Query 128 RCRP 131
RP
Sbjct 224 ETRP 227
> At5g50320
Length=559
Score = 67.8 bits (164), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 36/119 (30%), Positives = 66/119 (55%), Gaps = 12/119 (10%)
Query 13 CHYCPNEPGQ-----PRSYLSTEPAVLRANQNGWSPLKQFKDRAETLRRNGHVVDKIEVL 67
C YCP P +SY EP +RA + ++P Q + R + L+R GH VDK+E +
Sbjct 121 CVYCPGGPDSDFEYSTQSYTGYEPTSMRAIRARYNPYVQARSRIDQLKRLGHSVDKVEFI 180
Query 68 VLGGTWSGYPQDYQEEFIRDVYYAANVFGAPEPVRQPLSLEEEQSLNETADCRIVGLTL 126
++GGT+ P +Y++ FIR+++ A + + ++EE + +E + + +G+T+
Sbjct 181 LMGGTFMSLPAEYRDFFIRNLHDALSGHTSA-------NVEEAVAYSEHSATKCIGMTI 232
> 7295707
Length=552
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 36/119 (30%), Positives = 65/119 (54%), Gaps = 12/119 (10%)
Query 13 CHYCPNEPGQ-----PRSYLSTEPAVLRANQNGWSPLKQFKDRAETLRRNGHVVDKIEVL 67
C YCP P +SY EP +RA ++ + P Q + R E L++ GH VDK+E +
Sbjct 111 CVYCPGGPDSDFEYSTQSYTGYEPTSMRAIRSRYDPFLQTRHRVEQLKQLGHSVDKVEFI 170
Query 68 VLGGTWSGYPQDYQEEFIRDVYYAANVFGAPEPVRQPLSLEEEQSLNETADCRIVGLTL 126
V+GGT+ P++Y++ FIR+++ A + + ++ E +E + + +G+T+
Sbjct 171 VMGGTFMCLPEEYRDYFIRNLHDALSGHSSA-------NVAEAVRYSEKSRTKCIGITI 222
> Hs21361689
Length=547
Score = 67.0 bits (162), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 32/84 (38%), Positives = 50/84 (59%), Gaps = 5/84 (5%)
Query 13 CHYCPNEPGQ-----PRSYLSTEPAVLRANQNGWSPLKQFKDRAETLRRNGHVVDKIEVL 67
C YCP P +SY EP +RA + + P Q + R E L++ GH VDK+E +
Sbjct 109 CVYCPGGPDSDFEYSTQSYTGYEPTSMRAIRARYDPFLQTRHRIEQLKQLGHSVDKVEFI 168
Query 68 VLGGTWSGYPQDYQEEFIRDVYYA 91
V+GGT+ P++Y++ FIR+++ A
Sbjct 169 VMGGTFMALPEEYRDYFIRNLHDA 192
> HsM8922418
Length=499
Score = 67.0 bits (162), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 32/84 (38%), Positives = 50/84 (59%), Gaps = 5/84 (5%)
Query 13 CHYCPNEPGQ-----PRSYLSTEPAVLRANQNGWSPLKQFKDRAETLRRNGHVVDKIEVL 67
C YCP P +SY EP +RA + + P Q + R E L++ GH VDK+E +
Sbjct 109 CVYCPGGPDSDFEYSTQSYTGYEPTSMRAIRARYDPFLQTRHRIEQLKQLGHSVDKVEFI 168
Query 68 VLGGTWSGYPQDYQEEFIRDVYYA 91
V+GGT+ P++Y++ FIR+++ A
Sbjct 169 VMGGTFMALPEEYRDYFIRNLHDA 192
> YPL086c
Length=557
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 35/119 (29%), Positives = 65/119 (54%), Gaps = 12/119 (10%)
Query 13 CHYCPNEPGQ-----PRSYLSTEPAVLRANQNGWSPLKQFKDRAETLRRNGHVVDKIEVL 67
C YCP P +SY EP +RA + + P +Q + R E L++ GH +DK+E +
Sbjct 118 CVYCPGGPDSDFEYSTQSYTGYEPTSMRAIRARYDPYEQARGRVEQLKQLGHSIDKVEYV 177
Query 68 VLGGTWSGYPQDYQEEFIRDVYYAANVFGAPEPVRQPLSLEEEQSLNETADCRIVGLTL 126
++GGT+ P++Y+E+FI ++ A + F + ++E ++ + + VG+T+
Sbjct 178 LMGGTFMSLPKEYREDFIVKLHNALSGFNGND-------IDEAILYSQQSLTKCVGITI 229
> ECU08g1090
Length=613
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 34/119 (28%), Positives = 61/119 (51%), Gaps = 12/119 (10%)
Query 13 CHYCPNEPGQ-----PRSYLSTEPAVLRANQNGWSPLKQFKDRAETLRRNGHVVDKIEVL 67
C YCP P +SY EP +RA ++ + P +Q ++R L+ GH DK+E +
Sbjct 150 CVYCPGGPDSDFEYSTQSYTGYEPTSMRAIRSRYDPFEQARERLRQLKALGHNTDKVEYI 209
Query 68 VLGGTWSGYPQDYQEEFIRDVYYAANVFGAPEPVRQPLSLEEEQSLNETADCRIVGLTL 126
++GGT+ P+ Y++ FI ++ A + +P ++EE + + VG+T+
Sbjct 210 IMGGTFMSLPESYRDRFISRLHDALSGHTSP-------TVEEAIRVGAEGRLKCVGITI 261
> CE24765
Length=747
Score = 32.3 bits (72), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 15/46 (32%), Positives = 23/46 (50%), Gaps = 0/46 (0%)
Query 54 LRRNGHVVDKIEVLVLGGTWSGYPQDYQEEFIRDVYYAANVFGAPE 99
+ N HV + ++LV G + +P DYQ E I + + G PE
Sbjct 6 FKSNVHVQEYKDLLVFGYASTIFPNDYQSEHIAEERHTVPCLGDPE 51
> At2g20800
Length=582
Score = 32.0 bits (71), Expect = 0.58, Method: Compositional matrix adjust.
Identities = 15/35 (42%), Positives = 22/35 (62%), Gaps = 0/35 (0%)
Query 42 SPLKQFKDRAETLRRNGHVVDKIEVLVLGGTWSGY 76
+PLK+ TL +G+ + K +V+VLG WSGY
Sbjct 42 NPLKRILHADATLDSDGNPIRKKKVVVLGSGWSGY 76
> CE19658
Length=634
Score = 28.9 bits (63), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 16/53 (30%), Positives = 30/53 (56%), Gaps = 1/53 (1%)
Query 82 EEFIRDVYYAANVFGAPEPVRQPLSLEEEQSLNETADCRIVGLTLVRCR-PQR 133
E+F+ D N + A +P+R+ L+ + +NE A+ +G T++ R P+R
Sbjct 422 EKFVDDHSTPLNYYAAYQPIREFLANNDVIVINEGANTMDIGRTMMPSRLPKR 474
> CE02935
Length=634
Score = 28.9 bits (63), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 16/53 (30%), Positives = 30/53 (56%), Gaps = 1/53 (1%)
Query 82 EEFIRDVYYAANVFGAPEPVRQPLSLEEEQSLNETADCRIVGLTLVRCR-PQR 133
E+F+ D N + A +P+R+ L+ + +NE A+ +G T++ R P+R
Sbjct 422 EKFVDDHSTPLNYYAAYQPIREFLANNDVIVINEGANTMDIGRTMMPSRLPKR 474
> At4g25970
Length=628
Score = 28.5 bits (62), Expect = 5.4, Method: Composition-based stats.
Identities = 23/95 (24%), Positives = 45/95 (47%), Gaps = 12/95 (12%)
Query 34 LRANQNGWSPLKQFKDRAETLRRNGHVVDKIEVLVLGGTWSGYPQDYQEEFIRDVYYAAN 93
+ + QN S L+ FKD+ + + +D +V WS Y Q + E F+R++
Sbjct 361 VESAQNIPSFLEFFKDQI-NMAEVKYPLDHFKVRKGNSWWSFYLQTFNEFFVREL----- 414
Query 94 VFGAPEPVRQPLS-LEEEQSLNETADCRIVGLTLV 127
+P +P++ ++++ ADCR++ V
Sbjct 415 -----KPGARPIACMDQDDVAVSAADCRLMAFQSV 444
> Hs8922392
Length=620
Score = 28.1 bits (61), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 25/44 (56%), Gaps = 8/44 (18%)
Query 128 RCRPQRSSSAPKVMR-------RSFREFLIPLLCREKTASIVTD 164
R RP RS + P++M+ SF+ + PLLCRE A + +D
Sbjct 95 RWRP-RSFARPELMKILYNSLNDSFKRLIYPLLCREFRAKLTSD 137
> At3g20140
Length=510
Score = 28.1 bits (61), Expect = 8.7, Method: Composition-based stats.
Identities = 16/70 (22%), Positives = 34/70 (48%), Gaps = 5/70 (7%)
Query 84 FIRDVYYAANVFGAPEPVRQPLSLEEEQSLNETADCRIVGLTLVRCRPQRSSSAPKVMRR 143
F +++ +N F + + + +E E+ LNE D ++G+ L CR + + K+ R
Sbjct 245 FKKEIMDVSNSF---DELLERFLVEHEEKLNEDQDMDMMGVLLAACRDKNAEC--KITRN 299
Query 144 SFREFLIPLL 153
+ + L+
Sbjct 300 HIKSLFVDLV 309
Lambda K H
0.319 0.136 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2317343104
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40