bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0947_orf1
Length=238
Score E
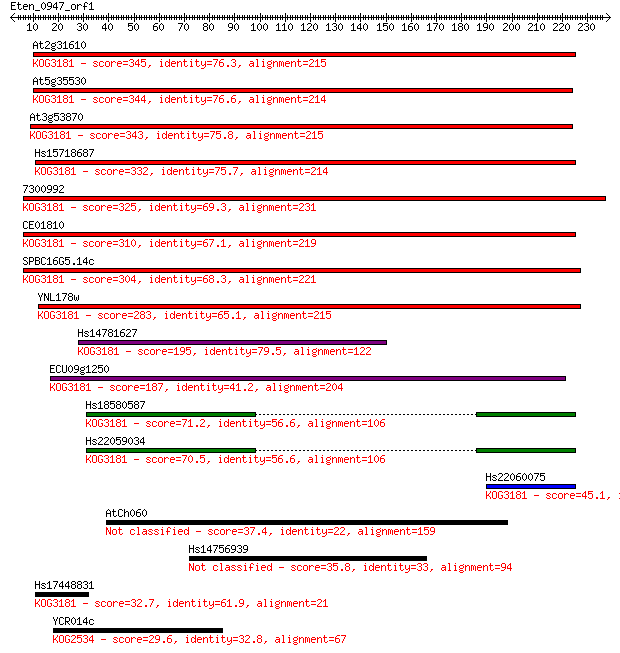
Sequences producing significant alignments: (Bits) Value
At2g31610 345 6e-95
At5g35530 344 9e-95
At3g53870 343 2e-94
Hs15718687 332 5e-91
7300992 325 4e-89
CE01810 310 2e-84
SPBC16G5.14c 304 8e-83
YNL178w 283 2e-76
Hs14781627 195 6e-50
ECU09g1250 187 1e-47
Hs18580587 71.2 2e-12
Hs22059034 70.5 3e-12
Hs22060075 45.1 1e-04
AtCh060 37.4 0.027
Hs14756939 35.8 0.080
Hs17448831 32.7 0.65
YCR014c 29.6 5.4
> At2g31610
Length=250
Score = 345 bits (884), Expect = 6e-95, Method: Compositional matrix adjust.
Identities = 164/215 (76%), Positives = 188/215 (87%), Gaps = 0/215 (0%)
Query 10 TNISKKRKFVNDGVFQAELNEFLSRTLAEDGYSGVEVRVTPIRTEIIIRATRTREVLGEK 69
T ISKKRKFV DGVF AELNE L+R LAEDGYSGVEVRVTP+RTEIIIRATRT+ VLGEK
Sbjct 3 TQISKKRKFVADGVFYAELNEVLTRELAEDGYSGVEVRVTPMRTEIIIRATRTQNVLGEK 62
Query 70 GRRIRELTSLVQKRFGFPPDSVELFAERVENRGLCAMAQAESLRYKLLKGLAVRRACYGV 129
GRRIRELTSLVQKRF FP DSVEL+AE+V NRGLCA+AQAESLRYKLL GLAVRRACYGV
Sbjct 63 GRRIRELTSLVQKRFKFPVDSVELYAEKVNNRGLCAIAQAESLRYKLLGGLAVRRACYGV 122
Query 130 LRHIMESGAKGCEVVVSGKLRAQRAKSMKFRDGYIISTGEPRQRFVSQAIRSVQLRQGVL 189
LR +MESGAKGCEV+VSGKLRA RAKSMKF+DGY++S+G+P + ++ A+R V LRQGVL
Sbjct 123 LRFVMESGAKGCEVIVSGKLRAARAKSMKFKDGYMVSSGQPTKEYIDAAVRHVLLRQGVL 182
Query 190 GVRVSIMLPFDPEGKMGPSTPLPDTIIVSEPKPEL 224
G++V IML +DP GK GP TPLPD +I+ PK ++
Sbjct 183 GIKVKIMLDWDPTGKSGPKTPLPDVVIIHAPKDDV 217
> At5g35530
Length=248
Score = 344 bits (883), Expect = 9e-95, Method: Compositional matrix adjust.
Identities = 164/214 (76%), Positives = 188/214 (87%), Gaps = 0/214 (0%)
Query 10 TNISKKRKFVNDGVFQAELNEFLSRTLAEDGYSGVEVRVTPIRTEIIIRATRTREVLGEK 69
T ISKKRKFV DGVF AELNE L+R LAEDGYSGVEVRVTP+RTEIIIRATRT+ VLGEK
Sbjct 3 TQISKKRKFVADGVFYAELNEVLTRELAEDGYSGVEVRVTPMRTEIIIRATRTQNVLGEK 62
Query 70 GRRIRELTSLVQKRFGFPPDSVELFAERVENRGLCAMAQAESLRYKLLKGLAVRRACYGV 129
GRRIRELTSLVQKRF FP DSVEL+AE+V NRGLCA+AQAESLRYKLL GLAVRRACYGV
Sbjct 63 GRRIRELTSLVQKRFKFPQDSVELYAEKVANRGLCAIAQAESLRYKLLGGLAVRRACYGV 122
Query 130 LRHIMESGAKGCEVVVSGKLRAQRAKSMKFRDGYIISTGEPRQRFVSQAIRSVQLRQGVL 189
LR +MESGAKGCEV+VSGKLRA RAKSMKF+DGY++S+G+P + ++ A+R V LRQGVL
Sbjct 123 LRFVMESGAKGCEVIVSGKLRAARAKSMKFKDGYMVSSGQPTKEYIDAAVRHVLLRQGVL 182
Query 190 GVRVSIMLPFDPEGKMGPSTPLPDTIIVSEPKPE 223
G++V IML +DP+GK GP TPLPD +I+ PK +
Sbjct 183 GLKVKIMLDWDPKGKQGPMTPLPDVVIIHTPKED 216
> At3g53870
Length=249
Score = 343 bits (880), Expect = 2e-94, Method: Compositional matrix adjust.
Identities = 163/215 (75%), Positives = 187/215 (86%), Gaps = 0/215 (0%)
Query 9 NTNISKKRKFVNDGVFQAELNEFLSRTLAEDGYSGVEVRVTPIRTEIIIRATRTREVLGE 68
T ISKKRKFV DGVF AELNE L+R LAEDGYSGVEVRVTP+RTEIIIRATRT+ VLGE
Sbjct 2 TTQISKKRKFVADGVFYAELNEVLTRELAEDGYSGVEVRVTPMRTEIIIRATRTQNVLGE 61
Query 69 KGRRIRELTSLVQKRFGFPPDSVELFAERVENRGLCAMAQAESLRYKLLKGLAVRRACYG 128
KGRRIRELTSLVQKRF FP DSVEL+AE+V NRGLCA+AQAESLRYKLL GLAVRRACYG
Sbjct 62 KGRRIRELTSLVQKRFKFPVDSVELYAEKVNNRGLCAIAQAESLRYKLLGGLAVRRACYG 121
Query 129 VLRHIMESGAKGCEVVVSGKLRAQRAKSMKFRDGYIISTGEPRQRFVSQAIRSVQLRQGV 188
VLR +MESGAKGCEV+VSGKLRA RAKSMKF+DGY++S+G+P + ++ A+R V LRQGV
Sbjct 122 VLRFVMESGAKGCEVIVSGKLRAARAKSMKFKDGYMVSSGQPTKEYIDSAVRHVLLRQGV 181
Query 189 LGVRVSIMLPFDPEGKMGPSTPLPDTIIVSEPKPE 223
LG++V +ML +DP+G GP TPLPD +I+ PK E
Sbjct 182 LGIKVKVMLDWDPKGISGPKTPLPDVVIIHSPKEE 216
> Hs15718687
Length=243
Score = 332 bits (850), Expect = 5e-91, Method: Compositional matrix adjust.
Identities = 162/214 (75%), Positives = 183/214 (85%), Gaps = 0/214 (0%)
Query 11 NISKKRKFVNDGVFQAELNEFLSRTLAEDGYSGVEVRVTPIRTEIIIRATRTREVLGEKG 70
ISKKRKFV DG+F+AELNEFL+R LAEDGYSGVEVRVTP RTEIII ATRT+ VLGEKG
Sbjct 4 QISKKRKFVADGIFKAELNEFLTRELAEDGYSGVEVRVTPTRTEIIILATRTQNVLGEKG 63
Query 71 RRIRELTSLVQKRFGFPPDSVELFAERVENRGLCAMAQAESLRYKLLKGLAVRRACYGVL 130
RRIRELT++VQKRFGFP SVEL+AE+V RGLCA+AQAESLRYKLL GLAVRRACYGVL
Sbjct 64 RRIRELTAVVQKRFGFPEGSVELYAEKVATRGLCAIAQAESLRYKLLGGLAVRRACYGVL 123
Query 131 RHIMESGAKGCEVVVSGKLRAQRAKSMKFRDGYIISTGEPRQRFVSQAIRSVQLRQGVLG 190
R IMESGAKGCEVVVSGKLR QRAKSMKF DG +I +G+P +V A+R V LRQGVLG
Sbjct 124 RFIMESGAKGCEVVVSGKLRGQRAKSMKFVDGLMIHSGDPVNYYVDTAVRHVLLRQGVLG 183
Query 191 VRVSIMLPFDPEGKMGPSTPLPDTIIVSEPKPEL 224
++V IMLP+DP GK+GP PLPD + + EPK E+
Sbjct 184 IKVKIMLPWDPTGKIGPKKPLPDHVSIVEPKDEI 217
> 7300992
Length=246
Score = 325 bits (834), Expect = 4e-89, Method: Compositional matrix adjust.
Identities = 160/231 (69%), Positives = 186/231 (80%), Gaps = 0/231 (0%)
Query 6 MAANTNISKKRKFVNDGVFQAELNEFLSRTLAEDGYSGVEVRVTPIRTEIIIRATRTREV 65
M AN ISKKRKFV+DG+F+AELNEFL+R LAEDGYSGVEVRVTP RTEIII AT+T++V
Sbjct 1 MNANLPISKKRKFVSDGIFKAELNEFLTRELAEDGYSGVEVRVTPSRTEIIIMATKTQQV 60
Query 66 LGEKGRRIRELTSLVQKRFGFPPDSVELFAERVENRGLCAMAQAESLRYKLLKGLAVRRA 125
LGEKGRRIRELT++VQKRF F +EL+AE+V RGLCA+AQAESLRYKL GLAVRRA
Sbjct 61 LGEKGRRIRELTAMVQKRFNFETGRIELYAEKVAARGLCAIAQAESLRYKLTGGLAVRRA 120
Query 126 CYGVLRHIMESGAKGCEVVVSGKLRAQRAKSMKFRDGYIISTGEPRQRFVSQAIRSVQLR 185
CYGVLR+IMESGAKGCEVVVSGKLR QRAKSMKF DG +I +G+P +V A R V LR
Sbjct 121 CYGVLRYIMESGAKGCEVVVSGKLRGQRAKSMKFVDGLMIHSGDPCNDYVETATRHVLLR 180
Query 186 QGVLGVRVSIMLPFDPEGKMGPSTPLPDTIIVSEPKPELPVAVQDPELAFP 236
QGVLG++V +MLP+DP+ K+GP PLPD + V EPK E + E P
Sbjct 181 QGVLGIKVKVMLPYDPKNKIGPKKPLPDNVSVVEPKEEKIYETPETEYKIP 231
> CE01810
Length=247
Score = 310 bits (794), Expect = 2e-84, Method: Compositional matrix adjust.
Identities = 147/219 (67%), Positives = 179/219 (81%), Gaps = 0/219 (0%)
Query 6 MAANTNISKKRKFVNDGVFQAELNEFLSRTLAEDGYSGVEVRVTPIRTEIIIRATRTREV 65
MAAN N++KK+K V G+F+AELN FL + LAEDGYSGVEVR TP R E+II ATRT+ V
Sbjct 1 MAANQNVTKKKKAVIGGIFKAELNNFLMKELAEDGYSGVEVRSTPARAEVIIMATRTQNV 60
Query 66 LGEKGRRIRELTSLVQKRFGFPPDSVELFAERVENRGLCAMAQAESLRYKLLKGLAVRRA 125
LGE+GRRI+ELTS+VQKRFGF SVEL+AE+V NRGLCA+AQ ESLRYKL+ GLAVRRA
Sbjct 61 LGERGRRIKELTSVVQKRFGFEEGSVELYAEKVSNRGLCAVAQCESLRYKLVGGLAVRRA 120
Query 126 CYGVLRHIMESGAKGCEVVVSGKLRAQRAKSMKFRDGYIISTGEPRQRFVSQAIRSVQLR 185
CYGVLR IMESGA+G EV+VSGKLR QRAK+MKF DG +I +G P ++ QA+R VQLR
Sbjct 121 CYGVLRFIMESGAQGVEVIVSGKLRGQRAKAMKFVDGLMIHSGHPVNDYIQQAVRHVQLR 180
Query 186 QGVLGVRVSIMLPFDPEGKMGPSTPLPDTIIVSEPKPEL 224
QGV+G++V IMLP+DP G+ GP LPD + + EP+ E+
Sbjct 181 QGVIGIKVKIMLPYDPRGQNGPRNALPDHVQIVEPQEEV 219
> SPBC16G5.14c
Length=249
Score = 304 bits (779), Expect = 8e-83, Method: Compositional matrix adjust.
Identities = 151/221 (68%), Positives = 182/221 (82%), Gaps = 3/221 (1%)
Query 6 MAANTNISKKRKFVNDGVFQAELNEFLSRTLAEDGYSGVEVRVTPIRTEIIIRATRTREV 65
M+A ISKKRKFV DGVF AELNEF +R L+E+GYSG EVRVTP R+EIIIRAT T++V
Sbjct 1 MSAAFTISKKRKFVADGVFYAELNEFFTRELSEEGYSGCEVRVTPSRSEIIIRATHTQDV 60
Query 66 LGEKGRRIRELTSLVQKRFGFPPDSVELFAERVENRGLCAMAQAESLRYKLLKGLAVRRA 125
LGEKGRRIRELT+LVQKRF F ++VEL+AE+V+NRGLCA+AQ ESLRYKLL GLAVRRA
Sbjct 61 LGEKGRRIRELTALVQKRFKFAENTVELYAEKVQNRGLCAVAQCESLRYKLLAGLAVRRA 120
Query 126 CYGVLRHIMESGAKGCEVVVSGKLRAQRAKSMKFRDGYIISTGEPRQRFVSQAIRSVQLR 185
YGVLR++ME+GAKGCEVV+SGKLRA RAKSMKF DG++I +G+P F+ A R V LR
Sbjct 121 AYGVLRYVMEAGAKGCEVVISGKLRAARAKSMKFADGFMIHSGQPAVDFIDSATRHVLLR 180
Query 186 QGVLGVRVSIMLPFDPEGKMGPSTPLPDTIIVSEPKPELPV 226
QGVLGV+V IMLP +P+ + S LPD ++V +PK E P+
Sbjct 181 QGVLGVKVKIMLP-EPKTRQKKS--LPDIVVVLDPKEEEPI 218
> YNL178w
Length=240
Score = 283 bits (725), Expect = 2e-76, Method: Compositional matrix adjust.
Identities = 140/216 (64%), Positives = 174/216 (80%), Gaps = 4/216 (1%)
Query 12 ISKKRKFVNDGVFQAELNEFLSRTLAEDGYSGVEVRVTPIRTEIIIRATRTREVLGEKGR 71
ISKKRK V DGVF AELNEF +R LAE+GYSGVEVRVTP +TE+IIRATRT++VLGE GR
Sbjct 5 ISKKRKLVADGVFYAELNEFFTRELAEEGYSGVEVRVTPTKTEVIIRATRTQDVLGENGR 64
Query 72 RIRELTSLVQKRFGFPPDSVELFAERVENRGLCAMAQAESLRYKLLKGLAVRRACYGVLR 131
RI ELT LVQKRF + P ++ L+AERV++RGL A+AQAES+++KLL GLA+RRA YGV+R
Sbjct 65 RINELTLLVQKRFKYAPGTIVLYAERVQDRGLSAVAQAESMKFKLLNGLAIRRAAYGVVR 124
Query 132 HIMESGAKGCEVVVSGKLRAQRAKSMKFRDGYIISTGEPRQRFVSQAIRSVQLRQGVLGV 191
++MESGAKGCEVVVSGKLRA RAK+MKF DG++I +G+P F+ A R V +RQGVLG+
Sbjct 125 YVMESGAKGCEVVVSGKLRAARAKAMKFADGFLIHSGQPVNDFIDTATRHVLMRQGVLGI 184
Query 192 RVSIMLPFDP-EGKMGPSTPLPDTIIVSEPKPELPV 226
+V IM DP + + GP LPD + + EPK E P+
Sbjct 185 KVKIMR--DPAKSRTGPKA-LPDAVTIIEPKEEEPI 217
> Hs14781627
Length=122
Score = 195 bits (496), Expect = 6e-50, Method: Compositional matrix adjust.
Identities = 97/122 (79%), Positives = 108/122 (88%), Gaps = 0/122 (0%)
Query 28 LNEFLSRTLAEDGYSGVEVRVTPIRTEIIIRATRTREVLGEKGRRIRELTSLVQKRFGFP 87
+NEFL++ LAEDGYSGVE+RVTP RTEII+ ATRT+ VLGEKGRRIRELT++VQKRFG P
Sbjct 1 MNEFLTQELAEDGYSGVEMRVTPTRTEIIVLATRTQNVLGEKGRRIRELTAVVQKRFGSP 60
Query 88 PDSVELFAERVENRGLCAMAQAESLRYKLLKGLAVRRACYGVLRHIMESGAKGCEVVVSG 147
SVEL+AE V RGLCA+AQAESL YKLL GLAVRRACYGV R IME+GAKGCEVVVSG
Sbjct 61 EGSVELYAEEVATRGLCAIAQAESLPYKLLGGLAVRRACYGVSRFIMETGAKGCEVVVSG 120
Query 148 KL 149
KL
Sbjct 121 KL 122
> ECU09g1250
Length=228
Score = 187 bits (476), Expect = 1e-47, Method: Compositional matrix adjust.
Identities = 84/204 (41%), Positives = 136/204 (66%), Gaps = 0/204 (0%)
Query 17 KFVNDGVFQAELNEFLSRTLAEDGYSGVEVRVTPIRTEIIIRATRTREVLGEKGRRIREL 76
+F+ +G+ AEL EF + L +G+S +E+R+ +II++ + E +GEK R+R+
Sbjct 21 RFMKNGLMNAELKEFFEKALVNEGFSTMELRMQETPIKIILKVAKPHEAIGEKKFRLRQF 80
Query 77 TSLVQKRFGFPPDSVELFAERVENRGLCAMAQAESLRYKLLKGLAVRRACYGVLRHIMES 136
L +R P +SVE+ E+V +GLCA+ QA +R K+L G+ RRA L+ +
Sbjct 81 QHLAAQRLEVPDESVEIVVEKVHEKGLCALIQANFIREKILGGVQYRRAVNMALKTARHA 140
Query 137 GAKGCEVVVSGKLRAQRAKSMKFRDGYIISTGEPRQRFVSQAIRSVQLRQGVLGVRVSIM 196
A+GC+++VSGKL+ QRAKS+KF+DG +I +G+ + +++ +V+ +QGV+G++V IM
Sbjct 141 KAQGCQIIVSGKLKGQRAKSVKFQDGVLIHSGDAVKDYINTGYATVETKQGVIGIQVRIM 200
Query 197 LPFDPEGKMGPSTPLPDTIIVSEP 220
LP+DPEG +GP+ PLPD I + EP
Sbjct 201 LPYDPEGVLGPNYPLPDRITILEP 224
> Hs18580587
Length=506
Score = 71.2 bits (173), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 37/67 (55%), Positives = 47/67 (70%), Gaps = 0/67 (0%)
Query 31 FLSRTLAEDGYSGVEVRVTPIRTEIIIRATRTREVLGEKGRRIRELTSLVQKRFGFPPDS 90
L TL +S + TP RTEIII ATRT+ VLGEKG+ + ELT+ V+KRFGFP S
Sbjct 360 LLYVTLTGYLFSFRSLSFTPTRTEIIILATRTQNVLGEKGQWVGELTAAVEKRFGFPEGS 419
Query 91 VELFAER 97
VEL+A++
Sbjct 420 VELYADK 426
Score = 58.2 bits (139), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 23/39 (58%), Positives = 30/39 (76%), Gaps = 0/39 (0%)
Query 186 QGVLGVRVSIMLPFDPEGKMGPSTPLPDTIIVSEPKPEL 224
+GVLG++V IMLP+DP GK GP PLPD + + EPK E+
Sbjct 426 KGVLGIKVKIMLPWDPTGKTGPKKPLPDHVSIVEPKDEI 464
> Hs22059034
Length=259
Score = 70.5 bits (171), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 37/67 (55%), Positives = 47/67 (70%), Gaps = 0/67 (0%)
Query 31 FLSRTLAEDGYSGVEVRVTPIRTEIIIRATRTREVLGEKGRRIRELTSLVQKRFGFPPDS 90
L TL +S + TP RTEIII ATRT+ VLGEKG+ + ELT+ V+KRFGFP S
Sbjct 113 LLYVTLTGYLFSFRSLSFTPTRTEIIILATRTQNVLGEKGQWVGELTAAVEKRFGFPEGS 172
Query 91 VELFAER 97
VEL+A++
Sbjct 173 VELYADK 179
Score = 57.0 bits (136), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 23/39 (58%), Positives = 30/39 (76%), Gaps = 0/39 (0%)
Query 186 QGVLGVRVSIMLPFDPEGKMGPSTPLPDTIIVSEPKPEL 224
+GVLG++V IMLP+DP GK GP PLPD + + EPK E+
Sbjct 179 KGVLGIKVKIMLPWDPTGKTGPKKPLPDHVSIVEPKDEI 217
> Hs22060075
Length=1069
Score = 45.1 bits (105), Expect = 1e-04, Method: Composition-based stats.
Identities = 18/35 (51%), Positives = 24/35 (68%), Gaps = 0/35 (0%)
Query 190 GVRVSIMLPFDPEGKMGPSTPLPDTIIVSEPKPEL 224
++V IMLP+ P GK GP PLPD + + EPK E+
Sbjct 661 AIKVKIMLPWGPAGKTGPKKPLPDHVSIVEPKDEI 695
> AtCh060
Length=218
Score = 37.4 bits (85), Expect = 0.027, Method: Compositional matrix adjust.
Identities = 35/161 (21%), Positives = 78/161 (48%), Gaps = 5/161 (3%)
Query 39 DGYSGVEVRVTPIRTEIIIRATRTREVLGEKGRRIRELTSLVQKRFGFPPDSVELFAERV 98
+G + +E++ +III + ++ +K RR+ EL VQK + + R+
Sbjct 58 EGIARIEIQKRIDLIQIIIYMGFPKLLIEDKPRRVEELQMNVQKELNCVNRKLNIAITRI 117
Query 99 ENRGLCAMAQAESLRYKLLKGLAVRRACYGVLRHIMESGAKGCEVVVSGKLRAQRAKSMK 158
N AE + +L ++ R+A + ++ KG +V ++G++ + ++
Sbjct 118 SNPYGDPNILAEFIAGQLKNRVSFRKAMKKAIELTEQANTKGIQVQIAGRIDGKEIARVE 177
Query 159 F-RDGYI-ISTGEPRQRFVSQAIRSVQLRQGVLGVRVSIML 197
+ R+G + + T E + + S +R++ GVLG+++ I +
Sbjct 178 WIREGRVPLQTIEAKIDYCSYTVRTI---YGVLGIKIWIFV 215
> Hs14756939
Length=1275
Score = 35.8 bits (81), Expect = 0.080, Method: Composition-based stats.
Identities = 31/95 (32%), Positives = 50/95 (52%), Gaps = 9/95 (9%)
Query 72 RIRELTSLVQKRFGFPPDSVELFAERVENRGL-CAMAQAESLRYKLLKGLAVRRACYGVL 130
++ EL +L + PP S L A +EN L C+ E LR + +K AV++ +L
Sbjct 1138 KLEELQTLCKT----PPRS--LSAGAIENACLPCSGGALEELRGQYIK--AVKKIKCDML 1189
Query 131 RHIMESGAKGCEVVVSGKLRAQRAKSMKFRDGYII 165
R+I ES + E+V + LR ++ + K R Y+I
Sbjct 1190 RYIQESKERAAEMVKAEVLRERQETARKMRKYYLI 1224
> Hs17448831
Length=256
Score = 32.7 bits (73), Expect = 0.65, Method: Compositional matrix adjust.
Identities = 13/21 (61%), Positives = 16/21 (76%), Gaps = 0/21 (0%)
Query 11 NISKKRKFVNDGVFQAELNEF 31
ISKKRKFV DG+ + EL +F
Sbjct 4 QISKKRKFVTDGILKVELKKF 24
> YCR014c
Length=582
Score = 29.6 bits (65), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 22/71 (30%), Positives = 37/71 (52%), Gaps = 5/71 (7%)
Query 18 FVNDGVFQAELNEFLSRTLAEDGYSGVEVRVTPIRTEI----IIRATRTREVLGEKGRRI 73
F ND A++ E L L +DGY +++TP ++ I+ RT +++G G R
Sbjct 376 FCNDTTELAKIMETLCIKLYKDGYIHCFLQLTPNLEKLFLKRIVERFRTAKIVG-YGERK 434
Query 74 RELTSLVQKRF 84
R +S + K+F
Sbjct 435 RWYSSEIIKKF 445
Lambda K H
0.320 0.137 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4740636838
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40