bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0960_orf2
Length=133
Score E
Sequences producing significant alignments: (Bits) Value
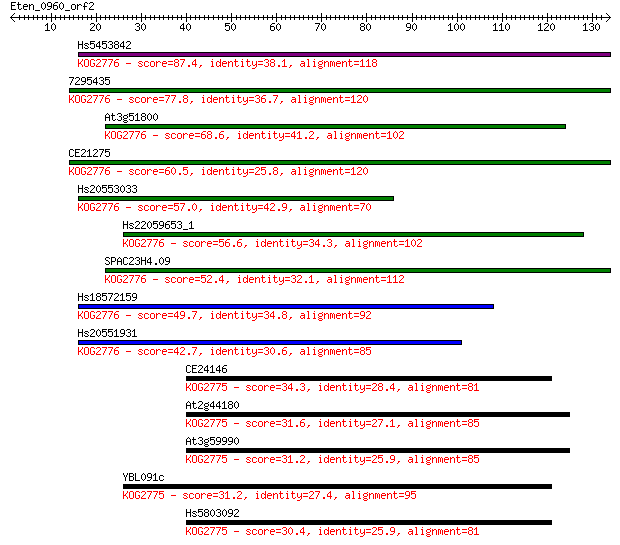
Hs5453842 87.4 7e-18
7295435 77.8 5e-15
At3g51800 68.6 3e-12
CE21275 60.5 9e-10
Hs20553033 57.0 9e-09
Hs22059653_1 56.6 1e-08
SPAC23H4.09 52.4 3e-07
Hs18572159 49.7 2e-06
Hs20551931 42.7 2e-04
CE24146 34.3 0.062
At2g44180 31.6 0.42
At3g59990 31.2 0.56
YBL091c 31.2 0.63
Hs5803092 30.4 0.89
> Hs5453842
Length=394
Score = 87.4 bits (215), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 45/121 (37%), Positives = 76/121 (62%), Gaps = 5/121 (4%)
Query 16 AAAAAAAGPAAAVLKACWLAAEAALRKVDVGAKASEVTKAIEAVAEDMGVKPMHGVLCYQ 75
A G A V+KA L AEAALR V G + ++VT+A VA P+ G+L +Q
Sbjct 130 AQGTQVTGRKADVIKAAHLCAEAALRLVKPGNQNTQVTEAWNKVAHSFNCTPIEGMLSHQ 189
Query 76 MKQHVIEGSRCFPIVSSSAEDKQEDF---TFEANELYSLDVVLSAGEGRARESAVRPSVF 132
+KQHVI+G + I+ + + +++D FE +E+Y++DV++S+GEG+A+++ R +++
Sbjct 190 LKQHVIDGEKT--IIQNPTDQQKKDHEKAEFEVHEVYAVDVLVSSGEGKAKDAGQRTTIY 247
Query 133 K 133
K
Sbjct 248 K 248
> 7295435
Length=391
Score = 77.8 bits (190), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 44/121 (36%), Positives = 71/121 (58%), Gaps = 1/121 (0%)
Query 14 AAAAAAAAAGPAAAVLKACWLAAEAALRKVDVGAKASEVTKAIEAVAEDMGVKPMHGVLC 73
AAA +G A V+ A + A +AALR + GA +T A++ ++E KP+ G+L
Sbjct 128 GAAADQKISGRQADVILAAYWAVQAALRLLKSGANNYSLTDAVQQISESYKCKPIEGMLS 187
Query 74 YQMKQHVIEGSRCFPIVSSSAEDKQ-EDFTFEANELYSLDVVLSAGEGRARESAVRPSVF 132
+++KQ I+G + S A+ K+ E TFE E+Y++DV++S GEG RE + S++
Sbjct 188 HELKQFKIDGEKTIIQNPSEAQRKEHEKCTFETYEVYAIDVIVSTGEGVGREKDTKVSIY 247
Query 133 K 133
K
Sbjct 248 K 248
> At3g51800
Length=392
Score = 68.6 bits (166), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 42/102 (41%), Positives = 65/102 (63%), Gaps = 1/102 (0%)
Query 22 AGPAAAVLKACWLAAEAALRKVDVGAKASEVTKAIEAVAEDMGVKPMHGVLCYQMKQHVI 81
+G A V+ A AA+ ALR V G K ++VT+AI+ VA K + GVL +Q+KQHVI
Sbjct 135 SGRKADVIAAANTAADVALRLVRPGKKNTDVTEAIQKVAAAYDCKIVEGVLSHQLKQHVI 194
Query 82 EGSRCFPIVSSSAEDKQEDFTFEANELYSLDVVLSAGEGRAR 123
+G++ + SS E ++ FE NE+Y++D+V S G+G+ +
Sbjct 195 DGNKVV-LSVSSPETTVDEVEFEENEVYAIDIVASTGDGKPK 235
> CE21275
Length=391
Score = 60.5 bits (145), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 31/123 (25%), Positives = 67/123 (54%), Gaps = 5/123 (4%)
Query 14 AAAAAAAAAGPAAAVLKACWLAAEAALRKVDVGAKASEVTKAIEAVAEDMGVKPMHGVLC 73
A+ G A +L+ A E A+R + + + +TK I+ A + G+ P+ +L
Sbjct 139 GASKDNKVTGKLADLLRGTHDALEIAIRSLRPDTENTTITKNIDKTAAEFGLTPIENMLS 198
Query 74 YQMKQHVIEGSRCFPIVSSSAEDKQ---EDFTFEANELYSLDVVLSAGEGRARESAVRPS 130
+Q++++ I+G + I+ +S E ++ E + +E Y++D++ S G+G+ ++ R +
Sbjct 199 HQLERNEIDGEK--KIIQNSGEKQKGEIEKIKIDKHEAYAIDILFSTGKGQPKDMDTRTT 256
Query 131 VFK 133
VF+
Sbjct 257 VFR 259
> Hs20553033
Length=210
Score = 57.0 bits (136), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 30/70 (42%), Positives = 41/70 (58%), Gaps = 0/70 (0%)
Query 16 AAAAAAAGPAAAVLKACWLAAEAALRKVDVGAKASEVTKAIEAVAEDMGVKPMHGVLCYQ 75
A G A V+KA L AEAALR V G + ++VT+A VA P+ G+L +Q
Sbjct 130 AQGTQVTGRKADVIKAAHLCAEAALRLVKPGNQNTQVTEAWNKVAHSFNCTPIEGMLSHQ 189
Query 76 MKQHVIEGSR 85
+KQHVI+G +
Sbjct 190 LKQHVIDGEK 199
> Hs22059653_1
Length=215
Score = 56.6 bits (135), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 35/102 (34%), Positives = 57/102 (55%), Gaps = 7/102 (6%)
Query 26 AAVLKACWLAAEAALRKVDVGAKASEVTKAIEAVAEDMGVKPMHGVLCYQMKQHVIEGSR 85
A V+KA L EAAL V G ++VT+A V P+ +L +Q+KQHVI+G +
Sbjct 105 ADVIKAAHLCVEAALCLVKPGNHNTQVTEAWNKVGHSFNCMPVEDMLSHQLKQHVIDGEK 164
Query 86 CFPIVSSSAEDKQEDFTFEANELYSLDVVLSAGEGRARESAV 127
V+ + D+Q+ + +E DV +S GEG+A+++ +
Sbjct 165 ---TVTQNPADQQK----KDHEKAEFDVFISIGEGKAKDAGL 199
> SPAC23H4.09
Length=381
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 36/114 (31%), Positives = 64/114 (56%), Gaps = 2/114 (1%)
Query 22 AGPAAAVLKACWLAAEAALRKVDVGAKASEVTKAIEAVAEDMGVKPMHGVLCYQMKQHVI 81
GPAA V+ A A +AA R + G +VT ++ +A G KP+ G+L +Q ++ VI
Sbjct 138 TGPAADVIAAASAALKAAQRTIKPGNTNWQVTDIVDKIATSYGCKPVAGMLSHQQEREVI 197
Query 82 EGSRCFPIVSSSAEDKQED-FTFEANELYSLDVVLSAG-EGRARESAVRPSVFK 133
+G + + S ++ + D FTFE E+Y +D+++S G+ + S + ++K
Sbjct 198 DGKKQVILNPSDSQRSEMDTFTFEEGEVYGVDILVSTSPSGKVKRSDIATRIYK 251
> Hs18572159
Length=910
Score = 49.7 bits (117), Expect = 2e-06, Method: Composition-based stats.
Identities = 32/93 (34%), Positives = 46/93 (49%), Gaps = 1/93 (1%)
Query 16 AAAAAAAGPAAAVLKACWLAAEAALRKVDVGAKASEVTKAIEAVAEDMGVKPMHGVLCYQ 75
A G A V+KA AEAA V G + ++VT+A VA P+ G+L +Q
Sbjct 538 AQGTQVTGRKADVIKAAHRCAEAAPCLVTPGNQNTQVTEAWNKVAHLFNCMPIEGMLSHQ 597
Query 76 MKQHVIEGSRCFPIVSSSAEDK-QEDFTFEANE 107
+KQHVI+G + + + + K E FE E
Sbjct 598 LKQHVIDGEKTITQIPTDKQKKDHEKAEFETAE 630
> Hs20551931
Length=307
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 26/85 (30%), Positives = 43/85 (50%), Gaps = 17/85 (20%)
Query 16 AAAAAAAGPAAAVLKACWLAAEAALRKVDVGAKASEVTKAIEAVAEDMGVKPMHGVLCYQ 75
A G A V+KA L AEAALR V G + ++ P+ G+L +Q
Sbjct 108 AQGTQVTGRKADVIKAAHLCAEAALRLVKPGNQNTQ---------------PIEGMLSHQ 152
Query 76 MKQHVIEGSRCFPIVSSSAEDKQED 100
+KQHVI+G + ++ + + +++D
Sbjct 153 LKQHVIDGEKT--VIQNPTDQQKKD 175
> CE24146
Length=444
Score = 34.3 bits (77), Expect = 0.062, Method: Composition-based stats.
Identities = 23/83 (27%), Positives = 40/83 (48%), Gaps = 7/83 (8%)
Query 40 LRKVDVGAKASEVTKA--IEAVAEDMGVKPMHGVLCYQMKQHVIEGSRCFPIVSSSAEDK 97
+R DVG EV + +E + VKP+ + + + Q+ I + PIV + K
Sbjct 259 VRLCDVGEIVEEVMTSHEVELDGKSYVVKPIRNLNGHSIAQYRIHAGKTVPIVKGGEQTK 318
Query 98 QEDFTFEANELYSLDVVLSAGEG 120
E+ NE+Y+++ S G+G
Sbjct 319 MEE-----NEIYAIETFGSTGKG 336
> At2g44180
Length=431
Score = 31.6 bits (70), Expect = 0.42, Method: Composition-based stats.
Identities = 23/87 (26%), Positives = 41/87 (47%), Gaps = 7/87 (8%)
Query 40 LRKVDVGAKASEVTKA--IEAVAEDMGVKPMHGVLCYQMKQHVIEGSRCFPIVSSSAEDK 97
+R DVGA EV ++ +E + VK + + + + ++ I + P V + K
Sbjct 248 VRLCDVGAAVQEVMESYEVEINGKVYQVKSIRNLNGHSIGRYQIHAEKSVPNVRGGEQTK 307
Query 98 QEDFTFEANELYSLDVVLSAGEGRARE 124
E+ ELY+++ S G+G RE
Sbjct 308 MEE-----GELYAIETFGSTGKGYVRE 329
> At3g59990
Length=435
Score = 31.2 bits (69), Expect = 0.56, Method: Composition-based stats.
Identities = 22/87 (25%), Positives = 40/87 (45%), Gaps = 7/87 (8%)
Query 40 LRKVDVGAKASEVTKA--IEAVAEDMGVKPMHGVLCYQMKQHVIEGSRCFPIVSSSAEDK 97
+R D+GA EV ++ +E + VK + + + + + I + PIV + K
Sbjct 252 VRLCDIGAAIQEVMESYEVEINGKVFQVKSIRNLNGHSIGPYQIHAGKSVPIVKGGEQTK 311
Query 98 QEDFTFEANELYSLDVVLSAGEGRARE 124
E+ E Y+++ S G+G RE
Sbjct 312 MEE-----GEFYAIETFGSTGKGYVRE 333
> YBL091c
Length=421
Score = 31.2 bits (69), Expect = 0.63, Method: Composition-based stats.
Identities = 26/100 (26%), Positives = 44/100 (44%), Gaps = 10/100 (10%)
Query 26 AAVLKACWLAAEAA---LRKVDVGAKASEVTKA--IEAVAEDMGVKPMHGVLCYQMKQHV 80
AAV A + + A +R D+G EV ++ +E E VKP + + + +
Sbjct 221 AAVKDATYTGIKEAGIDVRLTDIGEAIQEVMESYEVEINGETYQVKPCRNLCGHSIAPYR 280
Query 81 IEGSRCFPIVSSSAEDKQEDFTFEANELYSLDVVLSAGEG 120
I G + PIV + K E+ E ++++ S G G
Sbjct 281 IHGGKSVPIVKNGDTTKMEE-----GEHFAIETFGSTGRG 315
> Hs5803092
Length=478
Score = 30.4 bits (67), Expect = 0.89, Method: Composition-based stats.
Identities = 21/83 (25%), Positives = 39/83 (46%), Gaps = 7/83 (8%)
Query 40 LRKVDVGAKASEVTKA--IEAVAEDMGVKPMHGVLCYQMKQHVIEGSRCFPIVSSSAEDK 97
+R DVG EV ++ +E + VKP+ + + + Q+ I + PIV +
Sbjct 295 VRLCDVGEAIQEVMESYEVEIDGKTYQVKPIRNLNGHSIGQYRIHAGKTVPIVKGGEATR 354
Query 98 QEDFTFEANELYSLDVVLSAGEG 120
E+ E+Y+++ S G+G
Sbjct 355 MEE-----GEVYAIETFGSTGKG 372
Lambda K H
0.315 0.125 0.346
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1356426142
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40