bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0985_orf1
Length=240
Score E
Sequences producing significant alignments: (Bits) Value
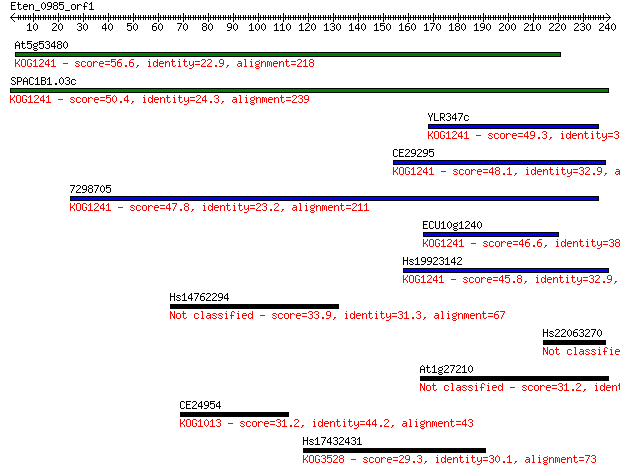
At5g53480 56.6 4e-08
SPAC1B1.03c 50.4 3e-06
YLR347c 49.3 6e-06
CE29295 48.1 2e-05
7298705 47.8 2e-05
ECU10g1240 46.6 5e-05
Hs19923142 45.8 7e-05
Hs14762294 33.9 0.28
Hs22063270 32.0 1.2
At1g27210 31.2 1.9
CE24954 31.2 2.1
Hs17432431 29.3 6.6
> At5g53480
Length=870
Score = 56.6 bits (135), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 50/218 (22%), Positives = 92/218 (42%), Gaps = 47/218 (21%)
Query 3 LLLLLDHFEGQLRKSFSFEQTEQTKQRQGMLCGVIQILCLRLGDQVKPVAGKLWNTLKQV 62
++ L + EG+ S ++ E+ + QG+LCG +Q++ +LG + P K Q+
Sbjct 553 MMELHNTLEGE---KLSLDEREKQNELQGLLCGCLQVIIQKLGSE--PTKSKFMEYADQM 607
Query 63 FQRPTHGLQVSQGESSAAPNNVHGFIVSSEPSVCDALLALSALINASGPAVSPFAGEVAD 122
GL + F S + +A+LA+ AL A+GP + + E
Sbjct 608 M-----GL------------FLRVFGCRSATAHEEAMLAIGALAYAAGPNFAKYMPEFYK 650
Query 123 LLVLQMQHQHEPQEAAASAVAATASTPTNDSSSADELQAARICIELVGDLSRALGTEFGS 182
L + +Q+ +E Q + + +VGD+ RAL +
Sbjct 651 YLEMGLQN-------------------------FEEYQVCAVTVGVVGDVCRALEDKILP 685
Query 183 LSDALLHGFYQLLRAPSVDRSLKPVVIVAVGDVALSLG 220
D ++ + L + + RS+KP + GD+AL++G
Sbjct 686 YCDGIMTQLLKDLSSNQLHRSVKPPIFSCFGDIALAIG 723
> SPAC1B1.03c
Length=863
Score = 50.4 bits (119), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 58/244 (23%), Positives = 98/244 (40%), Gaps = 55/244 (22%)
Query 1 NHLLLLLDHFEGQLR---KSFSFEQTEQTKQRQGMLCGVIQILCLRLGDQVKPVAGKLWN 57
N L ++L E ++ + E + Q LC V+ + R G ++ + ++ N
Sbjct 541 NVLSIILTRLETSIQMQSQILDVEDRANHDELQSNLCNVLTSIIRRFGPDIRTSSDQIMN 600
Query 58 TLKQVFQRPTHGLQVSQGESSAAPNN--VHGFIVSSEPSVCDALLALSALINASGPAVSP 115
L Q Q AP VH D LLA+ A++N+
Sbjct 601 LLLQTMQ--------------TAPKQSVVHE----------DVLLAIGAMMNS------- 629
Query 116 FAGEVADLLVLQMQHQHEPQEAAASAVAATASTPTNDSSSADELQAARICIELVGDLSRA 175
++ Q E S V +S +N+ E Q + + LVGDL+RA
Sbjct 630 ------------LEEQFEVY--VPSFVPFLSSALSNEQ----EYQLCSVAVGLVGDLARA 671
Query 176 LGTEFGSLSDALLHGFYQLLRAPSVDRSLKPVVIVAVGDVALSLGGAKFAPFTPWLIELL 235
L + D + Q L++ +DR++KP ++ D+AL++G A F + ++ LL
Sbjct 672 LNAKILPYCDDFMTRLVQDLQSSVLDRNVKPAILSCFSDIALAIGAA-FQTYLEAVMVLL 730
Query 236 VQAG 239
QA
Sbjct 731 QQAS 734
> YLR347c
Length=861
Score = 49.3 bits (116), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 22/68 (32%), Positives = 40/68 (58%), Gaps = 4/68 (5%)
Query 168 LVGDLSRALGTEFGSLSDALLHGFYQLLRAPSVDRSLKPVVIVAVGDVALSLGGAKFAPF 227
+ D+S +L +F SDA+++ Q++ P+ R LKP V+ GD+A ++G A F
Sbjct 662 FIADISNSLEEDFRRYSDAMMNVLAQMISNPNARRELKPAVLSVFGDIASNIG----ADF 717
Query 228 TPWLIELL 235
P+L +++
Sbjct 718 IPYLNDIM 725
> CE29295
Length=883
Score = 48.1 bits (113), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 28/85 (32%), Positives = 45/85 (52%), Gaps = 1/85 (1%)
Query 154 SSADELQAARICIELVGDLSRALGTEFGSLSDALLHGFYQLLRAPSVDRSLKPVVIVAVG 213
S+ DE Q + LV DLSRAL E D L+ L++P +DR++K V+I
Sbjct 656 SNTDETQVCAAAVGLVTDLSRALEAEIMPFMDELIQKLILCLQSPRLDRNVKVVIIGTFA 715
Query 214 DVALSLGGAKFAPFTPWLIELLVQA 238
D+A+++ A F + ++ +L A
Sbjct 716 DIAMAI-EAHFERYVGSVVPILNNA 739
> 7298705
Length=884
Score = 47.8 bits (112), Expect = 2e-05, Method: Composition-based stats.
Identities = 49/212 (23%), Positives = 87/212 (41%), Gaps = 48/212 (22%)
Query 25 QTKQRQGMLCGVIQILCLRLGDQVKP-VAGKLWNTLKQVFQRPTHGLQVSQGESSAAPNN 83
Q Q +LC +Q + ++ +Q P ++ + L +F S G+S
Sbjct 581 QFNDLQSLLCATLQSVLRKVHEQDAPQISDAIMTALLTMFNS-------SAGKSG----- 628
Query 84 VHGFIVSSEPSVCDALLALSALINASGPAVSPFAGEVADLLVLQMQHQHEPQEAAASAVA 143
+V E A LA+S L+ G + + D LV+ +++ E Q A+
Sbjct 629 ----VVQEE-----AFLAVSTLVELLGAQFAKYMPAFKDFLVMGLKNFQEYQVCCAA--- 676
Query 144 ATASTPTNDSSSADELQAARICIELVGDLSRALGTEFGSLSDALLHGFYQLLRAPSVDRS 203
+ L GD+ RAL S+ ++ L P++ R+
Sbjct 677 ----------------------VGLTGDIFRALKDLMVPYSNEIMTVLINNLTEPTIHRT 714
Query 204 LKPVVIVAVGDVALSLGGAKFAPFTPWLIELL 235
+KP V+ A GD+ALS+G F P+ ++++L
Sbjct 715 VKPQVLSAFGDIALSIGN-HFLPYLSMVLDML 745
> ECU10g1240
Length=854
Score = 46.6 bits (109), Expect = 5e-05, Method: Composition-based stats.
Identities = 21/54 (38%), Positives = 32/54 (59%), Gaps = 0/54 (0%)
Query 166 IELVGDLSRALGTEFGSLSDALLHGFYQLLRAPSVDRSLKPVVIVAVGDVALSL 219
I LVG L+ +GT+F L+ L Q L + + R LKP+++ GD+AL+L
Sbjct 671 INLVGRLANTMGTDFNILATVLTSSLVQCLSSEATHRDLKPIILSVFGDIALAL 724
> Hs19923142
Length=876
Score = 45.8 bits (107), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 27/82 (32%), Positives = 43/82 (52%), Gaps = 1/82 (1%)
Query 158 ELQAARICIELVGDLSRALGTEFGSLSDALLHGFYQLLRAPSVDRSLKPVVIVAVGDVAL 217
E Q + LVGDL RAL + D ++ + L +V RS+KP ++ GD+AL
Sbjct 663 EYQVCLAAVGLVGDLCRALQSNIIPFCDEVMQLLLENLGNENVHRSVKPQILSVFGDIAL 722
Query 218 SLGGAKFAPFTPWLIELLVQAG 239
++GG +F + ++ L QA
Sbjct 723 AIGG-EFKKYLEVVLNTLQQAS 743
> Hs14762294
Length=114
Score = 33.9 bits (76), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 21/67 (31%), Positives = 33/67 (49%), Gaps = 2/67 (2%)
Query 65 RPTHGLQVSQGESSAAPNNVHGFIVSSEPSVCDALLALSALINASGPAVSPFAGEVADLL 124
PT G + + SS + N+ H FI S S C L L+ L+ + + P A E +++
Sbjct 5 EPTEGQRTDEEVSSPSRNHHHSFIPSH--SQCPCLQPLNMLLFSPSSSQEPAAEETQEII 62
Query 125 VLQMQHQ 131
Q+ HQ
Sbjct 63 YAQLNHQ 69
> Hs22063270
Length=112
Score = 32.0 bits (71), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 12/25 (48%), Positives = 19/25 (76%), Gaps = 0/25 (0%)
Query 214 DVALSLGGAKFAPFTPWLIELLVQA 238
DV +L GA+ +P PWL+EL+++A
Sbjct 20 DVKPTLAGARLSPQQPWLLELMLRA 44
> At1g27210
Length=622
Score = 31.2 bits (69), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 33/75 (44%), Gaps = 2/75 (2%)
Query 165 CIELVGDLSRALGTEFGSLSDALLHGFYQLLRAPSVDRSLKPVVIVAVGDVALSLGGAKF 224
C+ L+ LSR G ++ + LR P D S++ VA D++ + F
Sbjct 93 CVALLSVLSRYHGDSLTPHLAKMVSTVIRRLRDP--DSSVRSACAVATADMSAHVTRQPF 150
Query 225 APFTPWLIELLVQAG 239
A LIE L+Q G
Sbjct 151 ASVAKPLIETLIQEG 165
> CE24954
Length=1069
Score = 31.2 bits (69), Expect = 2.1, Method: Composition-based stats.
Identities = 19/51 (37%), Positives = 25/51 (49%), Gaps = 8/51 (15%)
Query 69 GLQVSQGESSAAPNNV-----HG---FIVSSEPSVCDALLALSALINASGP 111
GLQ++ G +S PN HG F SS PS+C L A+ L + P
Sbjct 316 GLQMNGGPTSPLPNGTRRNTGHGGIEFPSSSRPSICSVLQAIEPLDRSKSP 366
> Hs17432431
Length=1205
Score = 29.3 bits (64), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 22/83 (26%), Positives = 37/83 (44%), Gaps = 16/83 (19%)
Query 118 GEVADLLVLQMQHQHEPQE----------AAASAVAATASTPTNDSSSADELQAARICIE 167
GE A L++ + + P+E + ++ T P NDS SA + +
Sbjct 459 GETASLVIARQEGHFLPRELKGEPDCCALSLETSEQLTFEIPLNDSGSAG------LGVS 512
Query 168 LVGDLSRALGTEFGSLSDALLHG 190
L G+ SR GT+ G +++HG
Sbjct 513 LKGNKSRETGTDLGIFIKSIIHG 535
Lambda K H
0.318 0.133 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4811924610
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40