bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
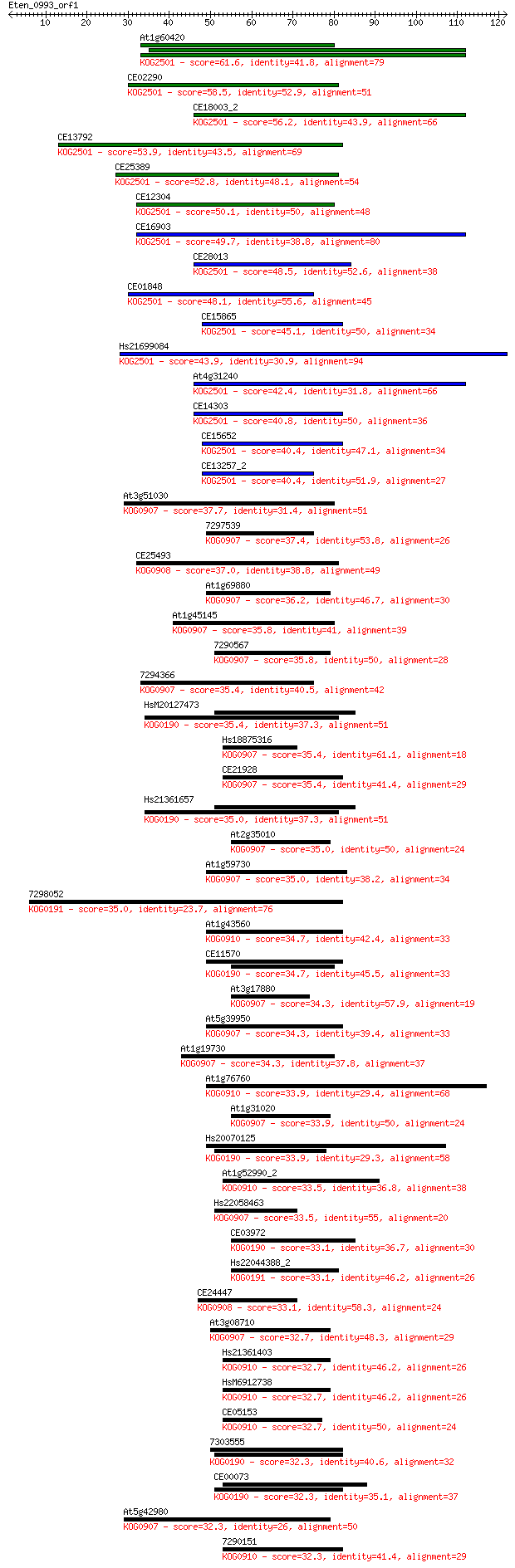
Query= Eten_0993_orf1
Length=121
Score E
Sequences producing significant alignments: (Bits) Value
At1g60420 61.6 3e-10
CE02290 58.5 3e-09
CE18003_2 56.2 1e-08
CE13792 53.9 8e-08
CE25389 52.8 2e-07
CE12304 50.1 1e-06
CE16903 49.7 1e-06
CE28013 48.5 3e-06
CE01848 48.1 4e-06
CE15865 45.1 4e-05
Hs21699084 43.9 7e-05
At4g31240 42.4 2e-04
CE14303 40.8 6e-04
CE15652 40.4 7e-04
CE13257_2 40.4 8e-04
At3g51030 37.7 0.006
7297539 37.4 0.008
CE25493 37.0 0.009
At1g69880 36.2 0.016
At1g45145 35.8 0.019
7290567 35.8 0.020
7294366 35.4 0.024
HsM20127473 35.4 0.028
Hs18875316 35.4 0.028
CE21928 35.4 0.030
Hs21361657 35.0 0.030
At2g35010 35.0 0.032
At1g59730 35.0 0.037
7298052 35.0 0.038
At1g43560 34.7 0.045
CE11570 34.7 0.046
At3g17880 34.3 0.055
At5g39950 34.3 0.060
At1g19730 34.3 0.061
At1g76760 33.9 0.075
At1g31020 33.9 0.076
Hs20070125 33.9 0.084
At1g52990_2 33.5 0.097
Hs22058463 33.5 0.099
CE03972 33.1 0.12
Hs22044388_2 33.1 0.13
CE24447 33.1 0.13
At3g08710 32.7 0.16
Hs21361403 32.7 0.17
HsM6912738 32.7 0.17
CE05153 32.7 0.18
7303555 32.3 0.20
CE00073 32.3 0.24
At5g42980 32.3 0.24
7290151 32.3 0.25
> At1g60420
Length=578
Score = 61.6 bits (148), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 26/47 (55%), Positives = 33/47 (70%), Gaps = 0/47 (0%)
Query 33 DKLIDNKGEAVDPSSLAGKTVGIYFSASWCGPCRNFTPKLSETVKKL 79
D L+ N GE V SL GK +G+YFSA+WCGPC+ FTP+L E +L
Sbjct 26 DFLVRNDGEQVKVDSLLGKKIGLYFSAAWCGPCQRFTPQLVEVYNEL 72
Score = 57.8 bits (138), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 30/79 (37%), Positives = 41/79 (51%), Gaps = 2/79 (2%)
Query 35 LIDNKGEAVDPSSLAGKTVGIYFSASWCGPCRNFTPKLSETVKKLKEGGNRWR--WFSSR 92
++ G V S L GKT+ +YFSA WC PCR FTPKL E K++KE + + SS
Sbjct 348 VLGKDGAKVLVSDLVGKTILMYFSAHWCPPCRAFTPKLVEVYKQIKERNEAFELIFISSD 407
Query 93 MTETKNHSKIISVKWIGLP 111
+ + W+ LP
Sbjct 408 RDQESFDEYYSQMPWLALP 426
Score = 40.4 bits (93), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 27/81 (33%), Positives = 35/81 (43%), Gaps = 2/81 (2%)
Query 33 DKLIDNKGEAVDPSSLAGKTVGIYFSASWCGPCRNFTPKLSETVKKLKEGGNRWR--WFS 90
D +I G V S L GKT+G+ FS + C TPKL E KLKE + S
Sbjct 186 DFVISPDGNKVPVSELEGKTIGLLFSVASYRKCTELTPKLVEFYTKLKENKEDFEIVLIS 245
Query 91 SRMTETKNHSKIISVKWIGLP 111
E + + W+ LP
Sbjct 246 LEDDEESFNQDFKTKPWLALP 266
> CE02290
Length=149
Score = 58.5 bits (140), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 27/52 (51%), Positives = 34/52 (65%), Gaps = 1/52 (1%)
Query 30 LLGDKLIDNKGEAVDP-SSLAGKTVGIYFSASWCGPCRNFTPKLSETVKKLK 80
L G KLI+ E +D L GK VG+YFSASWC PCR FTPKL+ +++
Sbjct 5 LAGTKLINQDSEELDAGEHLKGKVVGLYFSASWCPPCRQFTPKLTRFFDEIR 56
> CE18003_2
Length=144
Score = 56.2 bits (134), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 29/68 (42%), Positives = 42/68 (61%), Gaps = 3/68 (4%)
Query 46 SSLAGKTVGIYFSASWCGPCRNFTPKLSETVKKLKEGGNRWRWFSSRMTET--KNHSKII 103
+LAGK VGIYFSA WCGPCRNFTP L + +++++ + SS +E+ KN+ +
Sbjct 23 EALAGKIVGIYFSAHWCGPCRNFTPVLKDFYEEVQDDF-EIVFASSDQSESDLKNYMEEC 81
Query 104 SVKWIGLP 111
W +P
Sbjct 82 HGNWYYIP 89
> CE13792
Length=155
Score = 53.9 bits (128), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 30/70 (42%), Positives = 40/70 (57%), Gaps = 1/70 (1%)
Query 13 MLADPLGFPWPKKDPLQLLGDKLIDNKGEAVDPS-SLAGKTVGIYFSASWCGPCRNFTPK 71
M L + +P+K+ L G KL VD + +LAGK VG YFSA WC PCR FTP
Sbjct 1 MFLFQLIYTFPEKNMDILAGMKLEKLDKSLVDATEALAGKLVGFYFSAHWCPPCRGFTPI 60
Query 72 LSETVKKLKE 81
L + +++ E
Sbjct 61 LKDFYEEVNE 70
> CE25389
Length=149
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 26/55 (47%), Positives = 35/55 (63%), Gaps = 1/55 (1%)
Query 27 PLQLLGDKLIDNKGEAV-DPSSLAGKTVGIYFSASWCGPCRNFTPKLSETVKKLK 80
P L G L + +G+ + L GK +G+YFSASWC PCR FTPKL E +++K
Sbjct 2 PQFLKGVTLENREGDELPAEEHLKGKIIGLYFSASWCPPCRAFTPKLKEFFEEIK 56
> CE12304
Length=174
Score = 50.1 bits (118), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 24/49 (48%), Positives = 29/49 (59%), Gaps = 1/49 (2%)
Query 32 GDKLIDNKGEAVDP-SSLAGKTVGIYFSASWCGPCRNFTPKLSETVKKL 79
G L G VD L GK V +YFSASWCGPCR FTP + E +++
Sbjct 8 GTSLRLKDGTMVDAGEHLKGKIVVLYFSASWCGPCRQFTPIMKELYQQI 56
> CE16903
Length=151
Score = 49.7 bits (117), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 31/85 (36%), Positives = 44/85 (51%), Gaps = 5/85 (5%)
Query 32 GDKLIDNKGEAVDP-SSLAGK-TVGIYFSASWCGPCRNFTPKLSETVKKLKEGGNRWR-W 88
G LI+ +G+ +D +L GK V +YFSA WCG CR FTPKL + LK G
Sbjct 7 GKPLINQEGKELDGGDALRGKKVVALYFSAMWCGSCRQFTPKLKRFYEALKAAGKEIEVV 66
Query 89 FSSRMTETKNHSKIISV--KWIGLP 111
SR E ++ + + W+ +P
Sbjct 67 LVSRDREAEDLLEYLGHGGDWVAIP 91
> CE28013
Length=179
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 20/38 (52%), Positives = 24/38 (63%), Gaps = 0/38 (0%)
Query 46 SSLAGKTVGIYFSASWCGPCRNFTPKLSETVKKLKEGG 83
+L GK V +YFSA WC PC+ FTPKL LK+ G
Sbjct 50 EALKGKVVALYFSAGWCPPCKQFTPKLVRFYHHLKKAG 87
> CE01848
Length=140
Score = 48.1 bits (113), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 25/46 (54%), Positives = 28/46 (60%), Gaps = 1/46 (2%)
Query 30 LLGDKLIDNKGEAVDPS-SLAGKTVGIYFSASWCGPCRNFTPKLSE 74
L G KL VD + +LAGK VG YFSA WC PCR FTP L +
Sbjct 4 LAGVKLEKRDKTLVDATEALAGKAVGFYFSAHWCPPCRGFTPILKD 49
> CE15865
Length=777
Score = 45.1 bits (105), Expect = 4e-05, Method: Composition-based stats.
Identities = 17/34 (50%), Positives = 25/34 (73%), Gaps = 0/34 (0%)
Query 48 LAGKTVGIYFSASWCGPCRNFTPKLSETVKKLKE 81
L GK VG+YFSA WC P R+FTP L++ ++++
Sbjct 657 LDGKVVGLYFSAHWCPPSRDFTPVLAQFYSQVED 690
> Hs21699084
Length=135
Score = 43.9 bits (102), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 29/103 (28%), Positives = 49/103 (47%), Gaps = 11/103 (10%)
Query 28 LQLLGDK-LIDNKGEAVDP-SSLAGKTVGIYFSASWCGPCRNFTPKLSETVKKLKEGGNR 85
+ +LG++ L+ KG V+ ++L K V +YF+A+ C P R+FTP L + L R
Sbjct 2 VDILGERHLVTCKGATVEAEAALQNKVVALYFAAARCAPSRDFTPLLCDFYTALVAEARR 61
Query 86 WRWFSSRMT-------ETKNHSKIISVKWIGLPCHSVEPPQER 121
F E + + + W+ LP H +P ++R
Sbjct 62 PAPFEVVFVSADGSCQEMLDFMRELHGAWLALPFH--DPYRQR 102
> At4g31240
Length=204
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 21/69 (30%), Positives = 36/69 (52%), Gaps = 3/69 (4%)
Query 46 SSLAGKTVGIYFSASWCGPCRNFTPKLSETVKKL---KEGGNRWRWFSSRMTETKNHSKI 102
S L GKT+G+YF A WC P R+FT +L + +L +G S+ + + +
Sbjct 11 SKLVGKTIGLYFGAHWCPPFRSFTSQLVDVYNELATTDKGSFEVILISTDRDSREFNINM 70
Query 103 ISVKWIGLP 111
++ W+ +P
Sbjct 71 TNMPWLAIP 79
> CE14303
Length=122
Score = 40.8 bits (94), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 18/36 (50%), Positives = 23/36 (63%), Gaps = 0/36 (0%)
Query 46 SSLAGKTVGIYFSASWCGPCRNFTPKLSETVKKLKE 81
+LAGK G YFSA WC PC FTP L + +K+ +
Sbjct 11 EALAGKIGGFYFSAHWCPPCCMFTPILKKFYEKVYD 46
> CE15652
Length=178
Score = 40.4 bits (93), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 16/34 (47%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 48 LAGKTVGIYFSASWCGPCRNFTPKLSETVKKLKE 81
KTV +YFSA WCG C+ TPK+ + +KE
Sbjct 49 FENKTVVVYFSAGWCGSCKFLTPKIKKFYNAVKE 82
> CE13257_2
Length=745
Score = 40.4 bits (93), Expect = 8e-04, Method: Composition-based stats.
Identities = 14/27 (51%), Positives = 20/27 (74%), Gaps = 0/27 (0%)
Query 48 LAGKTVGIYFSASWCGPCRNFTPKLSE 74
L GK +G+Y+S WC P R+FTP L++
Sbjct 625 LDGKVIGLYYSGYWCQPSRDFTPILAQ 651
> At3g51030
Length=114
Score = 37.7 bits (86), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 28/51 (54%), Gaps = 0/51 (0%)
Query 29 QLLGDKLIDNKGEAVDPSSLAGKTVGIYFSASWCGPCRNFTPKLSETVKKL 79
Q++ ++ E + ++ + V + F+ASWCGPCR P ++ KKL
Sbjct 7 QVIACHTVETWNEQLQKANESKTLVVVDFTASWCGPCRFIAPFFADLAKKL 57
> 7297539
Length=106
Score = 37.4 bits (85), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 14/26 (53%), Positives = 19/26 (73%), Gaps = 0/26 (0%)
Query 49 AGKTVGIYFSASWCGPCRNFTPKLSE 74
+GK V + F A+WCGPC+ +PKL E
Sbjct 19 SGKLVVLDFFATWCGPCKMISPKLVE 44
> CE25493
Length=304
Score = 37.0 bits (84), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 19/49 (38%), Positives = 25/49 (51%), Gaps = 5/49 (10%)
Query 32 GDKLIDNKGEAVDPSSLAGKTVGIYFSASWCGPCRNFTPKLSETVKKLK 80
GD D K A + GK V + F+ASWCGPC+ P S+ + K
Sbjct 8 GDSDFDRKFSAGN-----GKAVFVDFTASWCGPCQYIAPIFSDLANQYK 51
> At1g69880
Length=148
Score = 36.2 bits (82), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 14/30 (46%), Positives = 17/30 (56%), Gaps = 0/30 (0%)
Query 49 AGKTVGIYFSASWCGPCRNFTPKLSETVKK 78
K + I F+A WCGPC+ PKL E K
Sbjct 58 TNKLLVIEFTAKWCGPCKTLEPKLEELAAK 87
> At1g45145
Length=118
Score = 35.8 bits (81), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 16/39 (41%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 41 EAVDPSSLAGKTVGIYFSASWCGPCRNFTPKLSETVKKL 79
E V ++ + K + I F+ASWC PCR P +E KK
Sbjct 18 EKVKDANESKKLIVIDFTASWCPPCRFIAPVFAEMAKKF 56
> 7290567
Length=111
Score = 35.8 bits (81), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 14/28 (50%), Positives = 17/28 (60%), Gaps = 0/28 (0%)
Query 51 KTVGIYFSASWCGPCRNFTPKLSETVKK 78
K V I F A WCGPC+ PKL E ++
Sbjct 21 KLVVIDFYADWCGPCKIIAPKLDELAQQ 48
> 7294366
Length=139
Score = 35.4 bits (80), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 17/42 (40%), Positives = 22/42 (52%), Gaps = 8/42 (19%)
Query 33 DKLIDNKGEAVDPSSLAGKTVGIYFSASWCGPCRNFTPKLSE 74
DKLID+ G K V + F A+WCGPC P+L +
Sbjct 18 DKLIDDAG--------TNKYVLVEFFATWCGPCAMIGPRLEQ 51
> HsM20127473
Length=505
Score = 35.4 bits (80), Expect = 0.028, Method: Composition-based stats.
Identities = 16/34 (47%), Positives = 19/34 (55%), Gaps = 0/34 (0%)
Query 51 KTVGIYFSASWCGPCRNFTPKLSETVKKLKEGGN 84
K V I F A WCG C+N PK E +KL + N
Sbjct 395 KDVLIEFYAPWCGHCKNLEPKYKELGEKLSKDPN 428
Score = 27.7 bits (60), Expect = 4.9, Method: Composition-based stats.
Identities = 12/47 (25%), Positives = 22/47 (46%), Gaps = 0/47 (0%)
Query 34 KLIDNKGEAVDPSSLAGKTVGIYFSASWCGPCRNFTPKLSETVKKLK 80
+L D+ E+ + + + + F A WCG C+ P+ +LK
Sbjct 29 ELTDDNFESRISDTGSAGLMLVEFFAPWCGHCKRLAPEYEAAATRLK 75
> Hs18875316
Length=486
Score = 35.4 bits (80), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 11/18 (61%), Positives = 13/18 (72%), Gaps = 0/18 (0%)
Query 53 VGIYFSASWCGPCRNFTP 70
V + FSA+WCGPCR P
Sbjct 404 VAVDFSATWCGPCRTIRP 421
> CE21928
Length=107
Score = 35.4 bits (80), Expect = 0.030, Method: Compositional matrix adjust.
Identities = 12/29 (41%), Positives = 20/29 (68%), Gaps = 0/29 (0%)
Query 53 VGIYFSASWCGPCRNFTPKLSETVKKLKE 81
V ++F+ASWCGPC+ P++ E + K+
Sbjct 23 VILFFTASWCGPCQMIKPRVEELAAEHKD 51
> Hs21361657
Length=505
Score = 35.0 bits (79), Expect = 0.030, Method: Composition-based stats.
Identities = 16/34 (47%), Positives = 19/34 (55%), Gaps = 0/34 (0%)
Query 51 KTVGIYFSASWCGPCRNFTPKLSETVKKLKEGGN 84
K V I F A WCG C+N PK E +KL + N
Sbjct 395 KDVLIEFYAPWCGHCKNLEPKYKELGEKLSKDPN 428
Score = 27.7 bits (60), Expect = 5.4, Method: Composition-based stats.
Identities = 12/47 (25%), Positives = 22/47 (46%), Gaps = 0/47 (0%)
Query 34 KLIDNKGEAVDPSSLAGKTVGIYFSASWCGPCRNFTPKLSETVKKLK 80
+L D+ E+ + + + + F A WCG C+ P+ +LK
Sbjct 29 ELTDDNFESRISDTGSAGLMLVEFFAPWCGHCKRLAPEYEAAATRLK 75
> At2g35010
Length=194
Score = 35.0 bits (79), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 12/24 (50%), Positives = 17/24 (70%), Gaps = 0/24 (0%)
Query 55 IYFSASWCGPCRNFTPKLSETVKK 78
YF+A+WCGPCR +P + E K+
Sbjct 111 FYFTAAWCGPCRFISPVIVELSKQ 134
> At1g59730
Length=129
Score = 35.0 bits (79), Expect = 0.037, Method: Compositional matrix adjust.
Identities = 13/34 (38%), Positives = 19/34 (55%), Gaps = 0/34 (0%)
Query 49 AGKTVGIYFSASWCGPCRNFTPKLSETVKKLKEG 82
+ K + I F+A WCGPC+ P++ E K E
Sbjct 42 SNKLLVIDFTAVWCGPCKAMEPRVREIASKYSEA 75
> 7298052
Length=510
Score = 35.0 bits (79), Expect = 0.038, Method: Composition-based stats.
Identities = 18/79 (22%), Positives = 36/79 (45%), Gaps = 3/79 (3%)
Query 6 NPQAYSSMLADPLGFPWPKKDPLQLLGD---KLIDNKGEAVDPSSLAGKTVGIYFSASWC 62
N +A S + +P P PK + D +++ + +P+ K+ + F A WC
Sbjct 241 NKEALVSFMLNPNAKPTPKPKEPEWSADTNSEIVHLTSQGFEPALKDEKSALVMFYAPWC 300
Query 63 GPCRNFTPKLSETVKKLKE 81
G C+ P+ + ++K+
Sbjct 301 GHCKRMKPEYEKAALEMKQ 319
> At1g43560
Length=112
Score = 34.7 bits (78), Expect = 0.045, Method: Compositional matrix adjust.
Identities = 14/33 (42%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 49 AGKTVGIYFSASWCGPCRNFTPKLSETVKKLKE 81
+ K V + F A+WCGPC+ P L+E + LK+
Sbjct 21 SDKPVLVDFYATWCGPCQLMVPILNEVSETLKD 53
> CE11570
Length=488
Score = 34.7 bits (78), Expect = 0.046, Method: Composition-based stats.
Identities = 15/33 (45%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 49 AGKTVGIYFSASWCGPCRNFTPKLSETVKKLKE 81
A K V I F A WCG C++ PK E +KL +
Sbjct 379 ADKDVLIEFYAPWCGHCKSLAPKYEELAEKLNK 411
Score = 26.9 bits (58), Expect = 9.2, Method: Composition-based stats.
Identities = 9/25 (36%), Positives = 12/25 (48%), Gaps = 0/25 (0%)
Query 55 IYFSASWCGPCRNFTPKLSETVKKL 79
+ F A WCG C+ P+ KL
Sbjct 42 VKFYAPWCGHCKKIAPEYERAAPKL 66
> At3g17880
Length=123
Score = 34.3 bits (77), Expect = 0.055, Method: Compositional matrix adjust.
Identities = 11/19 (57%), Positives = 15/19 (78%), Gaps = 0/19 (0%)
Query 55 IYFSASWCGPCRNFTPKLS 73
+YF+A+WCGPCR +P S
Sbjct 40 LYFTATWCGPCRYMSPLYS 58
> At5g39950
Length=133
Score = 34.3 bits (77), Expect = 0.060, Method: Compositional matrix adjust.
Identities = 13/33 (39%), Positives = 18/33 (54%), Gaps = 0/33 (0%)
Query 49 AGKTVGIYFSASWCGPCRNFTPKLSETVKKLKE 81
+ K + + FSASWCGPCR P + K +
Sbjct 46 SNKLLVVDFSASWCGPCRMIEPAIHAMADKFND 78
> At1g19730
Length=119
Score = 34.3 bits (77), Expect = 0.061, Method: Compositional matrix adjust.
Identities = 14/37 (37%), Positives = 21/37 (56%), Gaps = 0/37 (0%)
Query 43 VDPSSLAGKTVGIYFSASWCGPCRNFTPKLSETVKKL 79
+D + + K + I F+ASWC PCR P ++ KK
Sbjct 21 LDKAKESNKLIVIDFTASWCPPCRMIAPIFNDLAKKF 57
> At1g76760
Length=151
Score = 33.9 bits (76), Expect = 0.075, Method: Compositional matrix adjust.
Identities = 20/73 (27%), Positives = 33/73 (45%), Gaps = 5/73 (6%)
Query 49 AGKTVGIYFSASWCGPCRNFTPKLSETVKKLKEGGNRWRWFSSRMTETKNHSKIISVKWI 108
+ K V + + A+WCGPC+ P L+E + LK+ + + + N KI ++
Sbjct 59 SDKPVLVDYYATWCGPCQFMVPILNEVSETLKDKIQVVKIDTEKYPSIANKYKIEALPTF 118
Query 109 -----GLPCHSVE 116
G PC E
Sbjct 119 ILFKDGEPCDRFE 131
> At1g31020
Length=153
Score = 33.9 bits (76), Expect = 0.076, Method: Compositional matrix adjust.
Identities = 12/24 (50%), Positives = 16/24 (66%), Gaps = 0/24 (0%)
Query 55 IYFSASWCGPCRNFTPKLSETVKK 78
YF+A+WCGPCR +P + E K
Sbjct 70 FYFTAAWCGPCRLISPVILELSNK 93
> Hs20070125
Length=508
Score = 33.9 bits (76), Expect = 0.084, Method: Composition-based stats.
Identities = 17/58 (29%), Positives = 28/58 (48%), Gaps = 0/58 (0%)
Query 49 AGKTVGIYFSASWCGPCRNFTPKLSETVKKLKEGGNRWRWFSSRMTETKNHSKIISVK 106
A K + + F A WCG C+ P+ ++ KLK G+ R TE + ++ V+
Sbjct 40 AHKYLLVEFYAPWCGHCKALAPEYAKAAGKLKAEGSEIRLAKVDATEESDLAQQYGVR 97
Score = 30.0 bits (66), Expect = 1.0, Method: Composition-based stats.
Identities = 14/30 (46%), Positives = 16/30 (53%), Gaps = 3/30 (10%)
Query 51 KTVGIYFSASWCGPCRNFTP---KLSETVK 77
K V + F A WCG C+ P KL ET K
Sbjct 386 KNVFVEFYAPWCGHCKQLAPIWDKLGETYK 415
> At1g52990_2
Length=112
Score = 33.5 bits (75), Expect = 0.097, Method: Compositional matrix adjust.
Identities = 14/38 (36%), Positives = 23/38 (60%), Gaps = 3/38 (7%)
Query 53 VGIYFSASWCGPCRNFTPKLSETVKKLKEGGNRWRWFS 90
V + F+A WCGPCR+ P L+ K E N +++++
Sbjct 29 VMVMFTARWCGPCRDMIPILN---KMDSEYKNEFKFYT 63
> Hs22058463
Length=112
Score = 33.5 bits (75), Expect = 0.099, Method: Compositional matrix adjust.
Identities = 11/20 (55%), Positives = 14/20 (70%), Gaps = 0/20 (0%)
Query 51 KTVGIYFSASWCGPCRNFTP 70
+ V + FSA+WCGPCR P
Sbjct 28 RLVAVDFSATWCGPCRTIRP 47
> CE03972
Length=493
Score = 33.1 bits (74), Expect = 0.12, Method: Composition-based stats.
Identities = 11/30 (36%), Positives = 19/30 (63%), Gaps = 0/30 (0%)
Query 55 IYFSASWCGPCRNFTPKLSETVKKLKEGGN 84
+ F A WCG C++ P+ ++ +LKE G+
Sbjct 45 VEFYAPWCGHCKSLAPEYAKAATQLKEEGS 74
> Hs22044388_2
Length=666
Score = 33.1 bits (74), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 12/26 (46%), Positives = 16/26 (61%), Gaps = 0/26 (0%)
Query 55 IYFSASWCGPCRNFTPKLSETVKKLK 80
I F A WCGPC+NF P+ + +K
Sbjct 566 IDFYAPWCGPCQNFAPEFELLARMIK 591
> CE24447
Length=284
Score = 33.1 bits (74), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 14/25 (56%), Positives = 18/25 (72%), Gaps = 1/25 (4%)
Query 47 SLAG-KTVGIYFSASWCGPCRNFTP 70
SLAG K+V + F+A WCGPC+ P
Sbjct 17 SLAGLKSVIVDFTAVWCGPCKMIAP 41
> At3g08710
Length=140
Score = 32.7 bits (73), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 14/29 (48%), Positives = 17/29 (58%), Gaps = 0/29 (0%)
Query 50 GKTVGIYFSASWCGPCRNFTPKLSETVKK 78
GK V FSA+WCGPC+ P E +K
Sbjct 45 GKIVVANFSATWCGPCKIVAPFFIELSEK 73
> Hs21361403
Length=166
Score = 32.7 bits (73), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 12/26 (46%), Positives = 16/26 (61%), Gaps = 0/26 (0%)
Query 53 VGIYFSASWCGPCRNFTPKLSETVKK 78
V + F A WCGPC+ P+L + V K
Sbjct 81 VVVDFHAQWCGPCKILGPRLEKMVAK 106
> HsM6912738
Length=166
Score = 32.7 bits (73), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 12/26 (46%), Positives = 16/26 (61%), Gaps = 0/26 (0%)
Query 53 VGIYFSASWCGPCRNFTPKLSETVKK 78
V + F A WCGPC+ P+L + V K
Sbjct 81 VVVDFHAQWCGPCKILGPRLEKMVAK 106
> CE05153
Length=119
Score = 32.7 bits (73), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 12/24 (50%), Positives = 15/24 (62%), Gaps = 0/24 (0%)
Query 53 VGIYFSASWCGPCRNFTPKLSETV 76
V + F A WCGPC+ P+L E V
Sbjct 33 VIVDFHAEWCGPCQALGPRLEEKV 56
> 7303555
Length=488
Score = 32.3 bits (72), Expect = 0.20, Method: Composition-based stats.
Identities = 13/32 (40%), Positives = 18/32 (56%), Gaps = 0/32 (0%)
Query 50 GKTVGIYFSASWCGPCRNFTPKLSETVKKLKE 81
GK I F A WCG C+ +P E +KL++
Sbjct 381 GKDTLIEFYAPWCGHCKKLSPIYEELAEKLQD 412
Score = 27.7 bits (60), Expect = 5.5, Method: Composition-based stats.
Identities = 9/31 (29%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 51 KTVGIYFSASWCGPCRNFTPKLSETVKKLKE 81
+T + F A WCG C+ P+ ++ + +K+
Sbjct 39 ETTLVMFYAPWCGHCKRLKPEYAKAAEIVKD 69
> CE00073
Length=664
Score = 32.3 bits (72), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 12/35 (34%), Positives = 20/35 (57%), Gaps = 0/35 (0%)
Query 53 VGIYFSASWCGPCRNFTPKLSETVKKLKEGGNRWR 87
V + F A WCG C+ P+ + +KLK G++ +
Sbjct 213 VLVEFYAPWCGHCKKLAPEYEKAAQKLKAQGSKVK 247
Score = 31.2 bits (69), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 14/31 (45%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 51 KTVGIYFSASWCGPCRNFTPKLSETVKKLKE 81
K V I F A WCG C++F K E + LK+
Sbjct 564 KDVLIEFYAPWCGHCKSFESKYVELAQALKK 594
> At5g42980
Length=118
Score = 32.3 bits (72), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 13/50 (26%), Positives = 28/50 (56%), Gaps = 0/50 (0%)
Query 29 QLLGDKLIDNKGEAVDPSSLAGKTVGIYFSASWCGPCRNFTPKLSETVKK 78
+++ +++ E + ++ + K + I F+A+WC PCR P ++ KK
Sbjct 6 EVIACHTVEDWTEKLKAANESKKLIVIDFTATWCPPCRFIAPVFADLAKK 55
> 7290151
Length=160
Score = 32.3 bits (72), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 12/29 (41%), Positives = 17/29 (58%), Gaps = 0/29 (0%)
Query 53 VGIYFSASWCGPCRNFTPKLSETVKKLKE 81
V + F A WC PC+ TPK+ E ++ E
Sbjct 70 VIVNFHAEWCDPCKILTPKMLELLENSNE 98
Lambda K H
0.316 0.135 0.440
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1198419392
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40