bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0994_orf1
Length=165
Score E
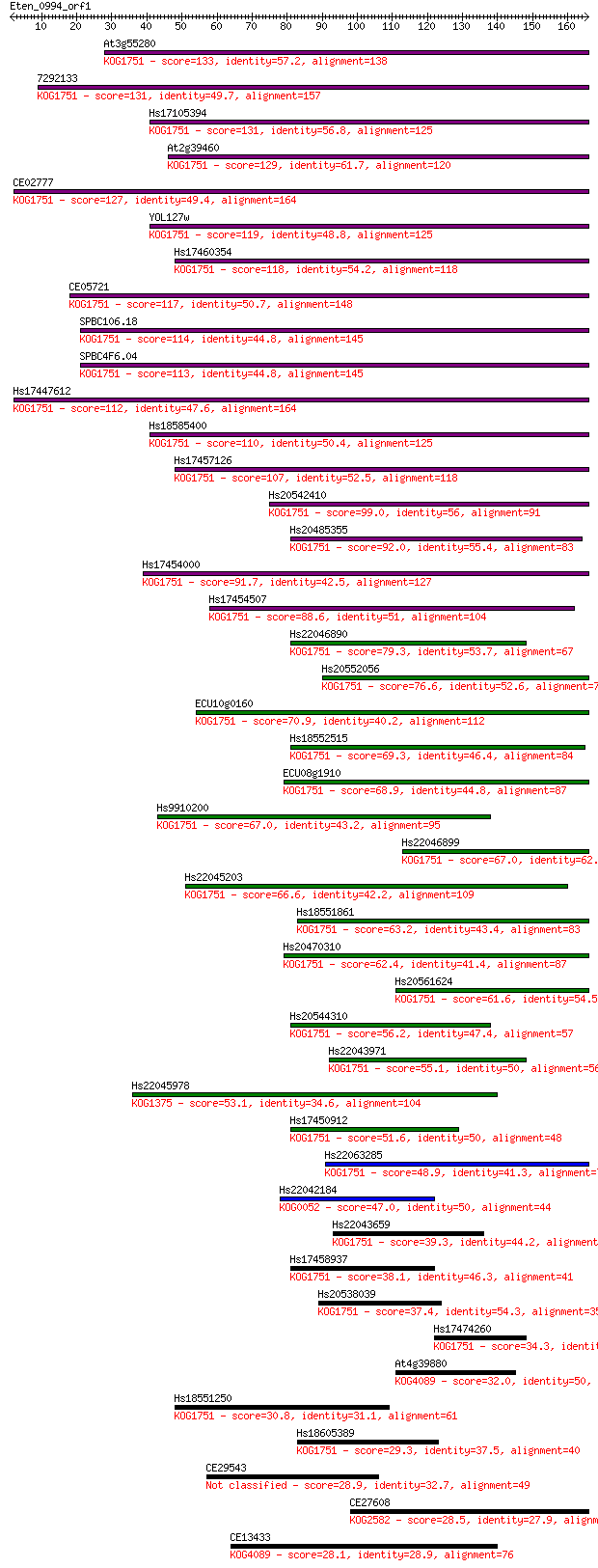
Sequences producing significant alignments: (Bits) Value
At3g55280 133 2e-31
7292133 131 6e-31
Hs17105394 131 7e-31
At2g39460 129 2e-30
CE02777 127 7e-30
YOL127w 119 2e-27
Hs17460354 118 7e-27
CE05721 117 1e-26
SPBC106.18 114 1e-25
SPBC4F6.04 113 1e-25
Hs17447612 112 2e-25
Hs18585400 110 1e-24
Hs17457126 107 7e-24
Hs20542410 99.0 3e-21
Hs20485355 92.0 5e-19
Hs17454000 91.7 6e-19
Hs17454507 88.6 5e-18
Hs22046890 79.3 3e-15
Hs20552056 76.6 2e-14
ECU10g0160 70.9 1e-12
Hs18552515 69.3 3e-12
ECU08g1910 68.9 5e-12
Hs9910200 67.0 1e-11
Hs22046899 67.0 2e-11
Hs22045203 66.6 2e-11
Hs18551861 63.2 3e-10
Hs20470310 62.4 4e-10
Hs20561624 61.6 6e-10
Hs20544310 56.2 3e-08
Hs22043971 55.1 5e-08
Hs22045978 53.1 3e-07
Hs17450912 51.6 7e-07
Hs22063285 48.9 5e-06
Hs22042184 47.0 2e-05
Hs22043659 39.3 0.003
Hs17458937 38.1 0.007
Hs20538039 37.4 0.014
Hs17474260 34.3 0.12
At4g39880 32.0 0.50
Hs18551250 30.8 1.3
Hs18605389 29.3 3.6
CE29543 28.9 4.7
CE27608 28.5 7.1
CE13433 28.1 7.4
> At3g55280
Length=154
Score = 133 bits (334), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 79/140 (56%), Positives = 101/140 (72%), Gaps = 15/140 (10%)
Query 28 GKALKKPGRTATAKARYTVRFRRPRTLKKPRTPKAPRLARLWAANPIAALQRN--DKYRV 85
G+ +KKP + K R V F RP+TL PR PK P+ I+A RN D Y++
Sbjct 28 GQIVKKPAK----KIRTKVTFHRPKTLTVPRKPKYPK---------ISATPRNKLDHYQI 74
Query 86 LQHPVTTESAMKKIEETNTLVFLCDPRASKRRIKEAVKETYGVVAQRVNTLIRVDGKKKA 145
L++P+TTESAMKKIE+ NTLVF+ D RA K++IK+AVK+ Y + ++VNTLIR DG KKA
Sbjct 75 LKYPLTTESAMKKIEDNNTLVFIVDIRADKKKIKDAVKKMYDIQTKKVNTLIRPDGTKKA 134
Query 146 YVRLTPEFDALDVANKIGII 165
YVRLTP++DALDVANKIGII
Sbjct 135 YVRLTPDYDALDVANKIGII 154
> 7292133
Length=277
Score = 131 bits (330), Expect = 6e-31, Method: Compositional matrix adjust.
Identities = 78/160 (48%), Positives = 101/160 (63%), Gaps = 12/160 (7%)
Query 9 PAKGS---KAAVEKKVKKAQAVGKALKKPGRTATAKARYTVRFRRPRTLKKPRTPKAPRL 65
PAKG+ KA KK Q K +K T K R V FRRP TLK PR+PK PR
Sbjct 127 PAKGTAKAKAVALLNAKKVQK--KIIKGAFGTRARKIRTNVHFRRPTTLKLPRSPKYPRK 184
Query 66 ARLWAANPIAALQRNDKYRVLQHPVTTESAMKKIEETNTLVFLCDPRASKRRIKEAVKET 125
+ + R D Y ++++P+TTE+AMKKIE+ NTLVFL RA+K ++ AV++
Sbjct 185 S-------VPTRNRMDAYNIIKYPLTTEAAMKKIEDNNTLVFLTHLRANKNHVRAAVRKL 237
Query 126 YGVVAQRVNTLIRVDGKKKAYVRLTPEFDALDVANKIGII 165
Y + +VN LIR DG+KKAYVRL ++DALD+ANKIGII
Sbjct 238 YDIKVAKVNVLIRPDGQKKAYVRLARDYDALDIANKIGII 277
> Hs17105394
Length=156
Score = 131 bits (329), Expect = 7e-31, Method: Compositional matrix adjust.
Identities = 71/127 (55%), Positives = 92/127 (72%), Gaps = 11/127 (8%)
Query 41 KARYTVRFRRPRTLKKPRTPKAPRLARLWAANPIAALQRN--DKYRVLQHPVTTESAMKK 98
K R + FRRP+TL+ R PK PR + A +RN D Y +++ P+TTESAMKK
Sbjct 39 KIRTSPTFRRPKTLRLRRQPKYPRKS---------APRRNKLDHYAIIKFPLTTESAMKK 89
Query 99 IEETNTLVFLCDPRASKRRIKEAVKETYGVVAQRVNTLIRVDGKKKAYVRLTPEFDALDV 158
IE+ NTLVF+ D +A+K +IK+AVK+ Y + +VNTLIR DG+KKAYVRL P++DALDV
Sbjct 90 IEDNNTLVFIVDVKANKHQIKQAVKKLYDIDVAKVNTLIRPDGEKKAYVRLAPDYDALDV 149
Query 159 ANKIGII 165
ANKIGII
Sbjct 150 ANKIGII 156
> At2g39460
Length=154
Score = 129 bits (324), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 74/122 (60%), Positives = 93/122 (76%), Gaps = 11/122 (9%)
Query 46 VRFRRPRTLKKPRTPKAPRLARLWAANPIAALQRN--DKYRVLQHPVTTESAMKKIEETN 103
V F RP+TL KPRT K P+ I+A RN D Y++L++P+TTESAMKKIE+ N
Sbjct 42 VTFHRPKTLTKPRTGKYPK---------ISATPRNKLDHYQILKYPLTTESAMKKIEDNN 92
Query 104 TLVFLCDPRASKRRIKEAVKETYGVVAQRVNTLIRVDGKKKAYVRLTPEFDALDVANKIG 163
TLVF+ D RA K++IK+AVK+ Y + ++VNTLIR DG KKAYVRLTP++DALDVANKIG
Sbjct 93 TLVFIVDIRADKKKIKDAVKKMYDIQTKKVNTLIRPDGTKKAYVRLTPDYDALDVANKIG 152
Query 164 II 165
II
Sbjct 153 II 154
> CE02777
Length=147
Score = 127 bits (320), Expect = 7e-30, Method: Compositional matrix adjust.
Identities = 81/164 (49%), Positives = 103/164 (62%), Gaps = 17/164 (10%)
Query 2 MAPKEKAPAKGSKAAVEKKVKKAQAVGKALKKPGRTATAKARYTVRFRRPRTLKKPRTPK 61
MAP APAK +KA KK K +K T + R +V FRRP TLK R +
Sbjct 1 MAPS--APAKTAKALDAKK--------KVVKGKRTTHRRQVRTSVHFRRPVTLKTARQAR 50
Query 62 APRLARLWAANPIAALQRNDKYRVLQHPVTTESAMKKIEETNTLVFLCDPRASKRRIKEA 121
PR + P + + D +R++QHP+TTESAMKKIEE NTLVF+ A+K +IK+A
Sbjct 51 FPR-----KSAPKTS--KMDHFRIIQHPLTTESAMKKIEEHNTLVFIVSNDANKYQIKDA 103
Query 122 VKETYGVVAQRVNTLIRVDGKKKAYVRLTPEFDALDVANKIGII 165
V + Y V A +VNTLI +KKAYVRLT ++DALDVANKIG+I
Sbjct 104 VHKLYNVQALKVNTLITPLQQKKAYVRLTADYDALDVANKIGVI 147
> YOL127w
Length=142
Score = 119 bits (299), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 61/125 (48%), Positives = 85/125 (68%), Gaps = 7/125 (5%)
Query 41 KARYTVRFRRPRTLKKPRTPKAPRLARLWAANPIAALQRNDKYRVLQHPVTTESAMKKIE 100
K R + FR P+TLK R PK +A+ + R D Y+V++ P+T+E+AMKK+E
Sbjct 25 KVRTSATFRLPKTLKLARAPK-------YASKAVPHYNRLDSYKVIEQPITSETAMKKVE 77
Query 101 ETNTLVFLCDPRASKRRIKEAVKETYGVVAQRVNTLIRVDGKKKAYVRLTPEFDALDVAN 160
+ N LVF +A+K +IK+AVKE Y V +VNTL+R +G KKAYVRLT ++DALD+AN
Sbjct 78 DGNILVFQVSMKANKYQIKKAVKELYEVDVLKVNTLVRPNGTKKAYVRLTADYDALDIAN 137
Query 161 KIGII 165
+IG I
Sbjct 138 RIGYI 142
> Hs17460354
Length=357
Score = 118 bits (295), Expect = 7e-27, Method: Compositional matrix adjust.
Identities = 64/120 (53%), Positives = 83/120 (69%), Gaps = 11/120 (9%)
Query 48 FRRPRTLKKPRTPKAPRLARLWAANPIAALQRN--DKYRVLQHPVTTESAMKKIEETNTL 105
FRRP+TL+ R PK P W + P +RN D Y +++ P+TTESAMKK + NT
Sbjct 247 FRRPKTLQLRRQPKYP-----WKSTP----RRNKLDHYAIIKFPLTTESAMKKTGDNNTR 297
Query 106 VFLCDPRASKRRIKEAVKETYGVVAQRVNTLIRVDGKKKAYVRLTPEFDALDVANKIGII 165
VF+ D +A+K +IK+AVK Y + +VNTLI DG+KKAYV L P++DALDVANKIGII
Sbjct 298 VFMVDVKANKHQIKQAVKRLYDIDMIKVNTLIWPDGEKKAYVPLAPDYDALDVANKIGII 357
> CE05721
Length=146
Score = 117 bits (293), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 75/149 (50%), Positives = 93/149 (62%), Gaps = 8/149 (5%)
Query 18 EKKVKKAQAVGKALKKPGRTATAK-ARYTVRFRRPRTLKKPRTPKAPRLARLWAANPIAA 76
KV KA KA+ K +T K R +V FRRP+TL T +APR AR A A
Sbjct 5 SNKVGKAIQAKKAVVKGSKTNVRKNVRTSVHFRRPKTL---VTARAPRYARKSAP----A 57
Query 77 LQRNDKYRVLQHPVTTESAMKKIEETNTLVFLCDPRASKRRIKEAVKETYGVVAQRVNTL 136
+ D + V++ P TTES+MKKIE+ NTLVF+ D +A+K IK AV Y V A +VNTL
Sbjct 58 RDKLDSFAVIKAPHTTESSMKKIEDHNTLVFIVDEKANKHHIKRAVHALYNVKAVKVNTL 117
Query 137 IRVDGKKKAYVRLTPEFDALDVANKIGII 165
I +KKAYVRL ++DALDVANKIG I
Sbjct 118 ITPLQQKKAYVRLASDYDALDVANKIGFI 146
> SPBC106.18
Length=141
Score = 114 bits (284), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 65/146 (44%), Positives = 94/146 (64%), Gaps = 8/146 (5%)
Query 21 VKKAQAVGKALKKPGRTATAK-ARYTVRFRRPRTLKKPRTPKAPRLARLWAANPIAALQR 79
V KA+ K ++K AK R + FRRP+TL+ R PK +A +A R
Sbjct 3 VAKAKGAQKTVQKGIHNKVAKKVRTSTTFRRPKTLQLSRKPK-------YARKSVAHAPR 55
Query 80 NDKYRVLQHPVTTESAMKKIEETNTLVFLCDPRASKRRIKEAVKETYGVVAQRVNTLIRV 139
D+Y+++ +P+ +ESAMKKIE+ NTLVF +A+K IKEAV++ Y V ++NTLIR
Sbjct 56 LDEYKIIVNPINSESAMKKIEDDNTLVFHVHLKANKFTIKEAVRKLYSVEPVKINTLIRP 115
Query 140 DGKKKAYVRLTPEFDALDVANKIGII 165
+G KKA+V+L+ + DALDVAN+IG +
Sbjct 116 NGTKKAFVKLSADADALDVANRIGFL 141
> SPBC4F6.04
Length=141
Score = 113 bits (283), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 65/146 (44%), Positives = 93/146 (63%), Gaps = 8/146 (5%)
Query 21 VKKAQAVGKALKKPGRTATA-KARYTVRFRRPRTLKKPRTPKAPRLARLWAANPIAALQR 79
V KA+ K ++K A K R + FRRP+TL+ R PK +A + R
Sbjct 3 VGKAKGAQKTVQKGIHNKVARKVRTSTTFRRPKTLELARKPK-------YARKSVPHASR 55
Query 80 NDKYRVLQHPVTTESAMKKIEETNTLVFLCDPRASKRRIKEAVKETYGVVAQRVNTLIRV 139
D+Y+++ +P+ +ESAMKKIE+ NTLVF +A+K IK AVK+ Y V A ++NTLIR
Sbjct 56 LDEYKIIVNPINSESAMKKIEDDNTLVFHVHLKANKFTIKNAVKKLYSVDAVKINTLIRP 115
Query 140 DGKKKAYVRLTPEFDALDVANKIGII 165
+G KKA+V+L+ + DALDVAN+IG +
Sbjct 116 NGTKKAFVKLSADADALDVANRIGFL 141
> Hs17447612
Length=157
Score = 112 bits (281), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 78/169 (46%), Positives = 102/169 (60%), Gaps = 17/169 (10%)
Query 2 MAPKEK--APAKGSKAAVEKKVKKAQAVGKALKKPGRTATAKARYTVRFRRPRTLKKPRT 59
MAPK K APA A +K +AV K + + K + FRRP+TL+ R
Sbjct 1 MAPKAKKEAPAPPKAEAKANALKAKKAVSKGVHSHTKN---KIHMSPTFRRPKTLRLRRQ 57
Query 60 PKAPRLARLWAANPIAALQRN--DKYRVLQHPVTTESAMKKIEETNTLVFLCDPRAS-KR 116
PK PR + A +RN D Y +++ P+TTE MKK EE NTLVF+ D +A+ K
Sbjct 58 PKYPRKS---------APRRNKLDHYAIIKFPLTTEFTMKKTEENNTLVFIVDVKATTKN 108
Query 117 RIKEAVKETYGVVAQRVNTLIRVDGKKKAYVRLTPEFDALDVANKIGII 165
+IK+A K+ Y + +VNT IR DG+KKAYVRL P ++ALDVANKIGII
Sbjct 109 QIKQAGKKLYDIDVAKVNTPIRPDGEKKAYVRLAPNYNALDVANKIGII 157
> Hs18585400
Length=156
Score = 110 bits (275), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 63/127 (49%), Positives = 86/127 (67%), Gaps = 11/127 (8%)
Query 41 KARYTVRFRRPRTLKKPRTPKAPRLARLWAANPIAALQRN--DKYRVLQHPVTTESAMKK 98
K R ++ F+RP+TL++ R P+ P W + P +RN Y V++ P+TTESA+K+
Sbjct 39 KTRTSLTFQRPKTLRRRRRPEYP-----WKSTP----RRNKLGHYAVIKFPLTTESAVKR 89
Query 99 IEETNTLVFLCDPRASKRRIKEAVKETYGVVAQRVNTLIRVDGKKKAYVRLTPEFDALDV 158
EE NTL+F D +A+K +IK+AVK+ Y V TLI DG+KKA VRL P++DALDV
Sbjct 90 TEENNTLLFTVDVKANKHQIKQAVKKLYDGDVAEVTTLIPPDGEKKACVRLAPDYDALDV 149
Query 159 ANKIGII 165
ANKIGII
Sbjct 150 ANKIGII 156
> Hs17457126
Length=155
Score = 107 bits (268), Expect = 7e-24, Method: Compositional matrix adjust.
Identities = 62/121 (51%), Positives = 84/121 (69%), Gaps = 12/121 (9%)
Query 48 FRRPRTLKKPRTPKAPRLARLWAANPIAALQRN--DKYRVLQHPVTTESAMKKIEETNT- 104
F+ P+TL+ R PK PR + P +RN D Y V++ +TTE AMKKIE+ N
Sbjct 44 FQWPKTLRLQRQPKDPR-----KSTP----RRNELDHYAVIKFLLTTEPAMKKIEDNNNM 94
Query 105 LVFLCDPRASKRRIKEAVKETYGVVAQRVNTLIRVDGKKKAYVRLTPEFDALDVANKIGI 164
LVF+ D +A+K +IK+AVK+ Y + + NTLIR DG+KKAYV+ +P++DALDVANKIGI
Sbjct 95 LVFIKDVKANKHQIKQAVKKLYNIDVAQFNTLIRPDGEKKAYVQPSPDYDALDVANKIGI 154
Query 165 I 165
I
Sbjct 155 I 155
> Hs20542410
Length=123
Score = 99.0 bits (245), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 51/93 (54%), Positives = 71/93 (76%), Gaps = 3/93 (3%)
Query 75 AALQRN--DKYRVLQHPVTTESAMKKIEETNTLVFLCDPRASKRRIKEAVKETYGVVAQR 132
+A +RN D Y V++ +TTESAMK IE+ N LVF+ D +A+K +IK+AVK+ Y + +
Sbjct 32 SAARRNNLDYYAVIKFLLTTESAMKNIEDNNMLVFIVDVKANKHQIKQAVKKLYDIDVAK 91
Query 133 VNTLIRVDGKKKAYVRLTPEFDALDVANKIGII 165
VNTLIR +G +KAYV+L P++DALDVANKIG +
Sbjct 92 VNTLIRPEG-EKAYVQLAPDYDALDVANKIGFM 123
> Hs20485355
Length=153
Score = 92.0 bits (227), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 46/83 (55%), Positives = 61/83 (73%), Gaps = 0/83 (0%)
Query 81 DKYRVLQHPVTTESAMKKIEETNTLVFLCDPRASKRRIKEAVKETYGVVAQRVNTLIRVD 140
D Y +++ P+TTESA+KK EE TLVF+ D +A+K +IK AVK+ Y +V TLI D
Sbjct 63 DHYAIIKFPLTTESAVKKTEENYTLVFIVDVKANKHQIKLAVKKLYDSDVAKVTTLICPD 122
Query 141 GKKKAYVRLTPEFDALDVANKIG 163
+KKAYVRL P++DALDVA K+G
Sbjct 123 KEKKAYVRLAPDYDALDVATKLG 145
> Hs17454000
Length=154
Score = 91.7 bits (226), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 54/127 (42%), Positives = 79/127 (62%), Gaps = 13/127 (10%)
Query 39 TAKARYTVRFRRPRTLKKPRTPKAPRLARLWAANPIAALQRNDKYRVLQHPVTTESAMKK 98
T K R + F RP+TL+ R K P+ + I + D Y +++ +TTESA +K
Sbjct 41 TKKDRTSPTFWRPKTLRLQRQQKYPQKS-------IPRRNKLDHYAIIKFLLTTESATEK 93
Query 99 IEETNTLVFLCDPRASKRRIKEAVKETYGVVAQRVNTLIRVDGKKKAYVRLTPEFDALDV 158
++ NTLVF D +A+K +IK+AVK+ Y + + DG+KKAYV+L P+++ALDV
Sbjct 94 TDDNNTLVFTVDAKANKHQIKQAVKKLYDI------DVAEPDGEKKAYVQLPPDYNALDV 147
Query 159 ANKIGII 165
ANKIGII
Sbjct 148 ANKIGII 154
> Hs17454507
Length=157
Score = 88.6 bits (218), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 53/106 (50%), Positives = 71/106 (66%), Gaps = 11/106 (10%)
Query 58 RTPKAPRLARLWAANPIAALQRN--DKYRVLQHPVTTESAMKKIEETNTLVFLCDPRASK 115
R PK PR + A +RN D+Y +++ TTESAMKKIE+ T VF D +A+K
Sbjct 55 RPPKYPRKS---------APRRNKLDRYAIIKFLWTTESAMKKIEDNTTYVFRVDVKANK 105
Query 116 RRIKEAVKETYGVVAQRVNTLIRVDGKKKAYVRLTPEFDALDVANK 161
++IK+AVK+ A +VNTLIR DG+KKAYV+L P++D LDVA K
Sbjct 106 QQIKQAVKKLPDNDAAKVNTLIRPDGEKKAYVQLAPDYDVLDVAKK 151
> Hs22046890
Length=159
Score = 79.3 bits (194), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 36/67 (53%), Positives = 53/67 (79%), Gaps = 0/67 (0%)
Query 81 DKYRVLQHPVTTESAMKKIEETNTLVFLCDPRASKRRIKEAVKETYGVVAQRVNTLIRVD 140
D Y +++ P+TTESA+KK E+ NTL+F+ D +A+K +IK+AVK+ Y + +VN+LIR D
Sbjct 93 DHYAIIELPLTTESAVKKTEDNNTLLFIVDVKANKHQIKQAVKKLYDMDVAKVNSLIRPD 152
Query 141 GKKKAYV 147
G+KKAYV
Sbjct 153 GEKKAYV 159
> Hs20552056
Length=93
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 40/76 (52%), Positives = 53/76 (69%), Gaps = 11/76 (14%)
Query 90 VTTESAMKKIEETNTLVFLCDPRASKRRIKEAVKETYGVVAQRVNTLIRVDGKKKAYVRL 149
+T ESAMKK E+ NTLVF+ D +A+K RIK+A+K+ Y +D KKAYV+L
Sbjct 29 LTAESAMKKTEDNNTLVFIVDVKANKHRIKQAMKKLY-----------DIDVAKKAYVQL 77
Query 150 TPEFDALDVANKIGII 165
P+ +AL+VANKIGII
Sbjct 78 APDINALEVANKIGII 93
> ECU10g0160
Length=129
Score = 70.9 bits (172), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 45/112 (40%), Positives = 64/112 (57%), Gaps = 2/112 (1%)
Query 54 LKKPRTPKAPRLARLWAANPIAALQRNDKYRVLQHPVTTESAMKKIEETNTLVFLCDPRA 113
L KP ++PR N + + RN ++ H +TTE A + +E+ NTLVF + +
Sbjct 20 LHKPSM-ESPRKNFKHPRNLVTKMPRNTPNDIILHALTTEKAARCMEQFNTLVFYVEKTS 78
Query 114 SKRRIKEAVKETYGVVAQRVNTLIRVDGKKKAYVRLTPEFDALDVANKIGII 165
+K IK+A+ E YG ++VNTLI G KKAYV E AL++A KIG I
Sbjct 79 TKPEIKKAITELYGSKVKKVNTLINKHG-KKAYVSFADEGAALEIAAKIGTI 129
> Hs18552515
Length=92
Score = 69.3 bits (168), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 39/84 (46%), Positives = 56/84 (66%), Gaps = 2/84 (2%)
Query 81 DKYRVLQHPVTTESAMKKIEETNTLVFLCDPRASKRRIKEAVKETYGVVAQRVNTLIRVD 140
D Y +++ P+TT+SAMK E NT+V + D +A+K +IK +K+ Y +V+TLIR
Sbjct 10 DHYAIIKFPLTTDSAMKS-EANNTVVLIEDVKANKHKIKLTMKKLYKTDVAKVHTLIR-P 67
Query 141 GKKKAYVRLTPEFDALDVANKIGI 164
K+ YV+L P+ D LDVANKI I
Sbjct 68 KKENTYVQLAPDNDCLDVANKIRI 91
> ECU08g1910
Length=88
Score = 68.9 bits (167), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 39/87 (44%), Positives = 54/87 (62%), Gaps = 1/87 (1%)
Query 79 RNDKYRVLQHPVTTESAMKKIEETNTLVFLCDPRASKRRIKEAVKETYGVVAQRVNTLIR 138
RN ++ H +TTE A + +E+ NTLVF + ++K IK+A+ E YG ++VNTLI
Sbjct 3 RNTPNDIILHALTTEKAARCMEQFNTLVFYVEKTSTKPEIKKAITELYGSKVKKVNTLIN 62
Query 139 VDGKKKAYVRLTPEFDALDVANKIGII 165
G KKAYV E AL++A KIG I
Sbjct 63 KHG-KKAYVSFADEGAALEIAAKIGTI 88
> Hs9910200
Length=130
Score = 67.0 bits (162), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 41/97 (42%), Positives = 61/97 (62%), Gaps = 11/97 (11%)
Query 43 RYTVRFRRPRTLKKPRTPKAPRLARLWAANPIAALQRN--DKYRVLQHPVTTESAMKKIE 100
R + F+RP+TL+ R P+ PR P +RN D Y +++ P+TTE AMKKI+
Sbjct 41 RMSPTFQRPKTLRLWRPPRYPR-----KTTP----RRNKLDHYAIIKFPLTTEFAMKKIK 91
Query 101 ETNTLVFLCDPRASKRRIKEAVKETYGVVAQRVNTLI 137
+ NTLVF D +A+K +IK+AVK+ + +VNTL+
Sbjct 92 DNNTLVFTVDVKANKHQIKQAVKKLCDIDGAKVNTLM 128
> Hs22046899
Length=458
Score = 67.0 bits (162), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 33/53 (62%), Positives = 41/53 (77%), Gaps = 0/53 (0%)
Query 113 ASKRRIKEAVKETYGVVAQRVNTLIRVDGKKKAYVRLTPEFDALDVANKIGII 165
A+K +IK AVK+ Y + +VNTLIR D +KKAYV L P++DALDVA KIGII
Sbjct 406 ANKHQIKPAVKKLYDMDVAKVNTLIRCDREKKAYVELPPDYDALDVAGKIGII 458
> Hs22045203
Length=112
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 46/109 (42%), Positives = 61/109 (55%), Gaps = 18/109 (16%)
Query 51 PRTLKKPRTPKAPRLARLWAANPIAALQRNDKYRVLQHPVTTESAMKKIEETNTLVFLCD 110
P L K + APR +L D Y V + P+TTESAMKKI++ NTLVF+
Sbjct 7 PLALGKNTSKSAPRRNKL------------DHYAV-KIPLTTESAMKKIDD-NTLVFIWL 52
Query 111 PRASKRRIKEAVKETYGVVAQRVNTLIRVDGKKKAYVRLTPEFDALDVA 159
+ + R + +VNTLI DG+KKAYVRL P+++ALDVA
Sbjct 53 LKTTSTRSNRLCD----IDVAKVNTLIHPDGEKKAYVRLAPDYNALDVA 97
> Hs18551861
Length=98
Score = 63.2 bits (152), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 36/83 (43%), Positives = 48/83 (57%), Gaps = 17/83 (20%)
Query 83 YRVLQHPVTTESAMKKIEETNTLVFLCDPRASKRRIKEAVKETYGVVAQRVNTLIRVDGK 142
Y ++ + TESAMKK+E+ NTLVF+ D +A+K +IKEA K
Sbjct 33 YAIIIFHLITESAMKKVEDNNTLVFIVDVKANKHQIKEATKNL----------------- 75
Query 143 KKAYVRLTPEFDALDVANKIGII 165
+ + LTP+ DA DVANKIGII
Sbjct 76 EDRDIPLTPDCDASDVANKIGII 98
> Hs20470310
Length=94
Score = 62.4 bits (150), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 36/87 (41%), Positives = 52/87 (59%), Gaps = 23/87 (26%)
Query 79 RNDKYRVLQHPVTTESAMKKIEETNTLVFLCDPRASKRRIKEAVKETYGVVAQRVNTLIR 138
R D Y +++ P+TTESAMK +E+ NTLV + D +A+ + +E +K
Sbjct 31 RLDHYAIIKFPLTTESAMK-MEDNNTLVLIVDVKANNTK-EEGIK--------------- 73
Query 139 VDGKKKAYVRLTPEFDALDVANKIGII 165
AYVRL P++DAL+VA+KIGII
Sbjct 74 ------AYVRLAPDYDALNVADKIGII 94
> Hs20561624
Length=124
Score = 61.6 bits (148), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 30/55 (54%), Positives = 41/55 (74%), Gaps = 0/55 (0%)
Query 111 PRASKRRIKEAVKETYGVVAQRVNTLIRVDGKKKAYVRLTPEFDALDVANKIGII 165
P K +I++AVK+ Y +V TLI+ DG+KKA+VRL ++DALDVANK+GII
Sbjct 70 PIFPKHQIEQAVKKLYDTDVAKVITLIQPDGEKKAHVRLASDYDALDVANKVGII 124
> Hs20544310
Length=340
Score = 56.2 bits (134), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 27/57 (47%), Positives = 39/57 (68%), Gaps = 0/57 (0%)
Query 81 DKYRVLQHPVTTESAMKKIEETNTLVFLCDPRASKRRIKEAVKETYGVVAQRVNTLI 137
D + + P+TT+S KKIE+ NTLVF+ D +A+K + K+AVK+ Y +VNTLI
Sbjct 190 DHCAITKFPLTTKSTTKKIEDNNTLVFIVDVKANKHQFKQAVKKLYDTDVAKVNTLI 246
> Hs22043971
Length=203
Score = 55.1 bits (131), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 28/56 (50%), Positives = 39/56 (69%), Gaps = 0/56 (0%)
Query 92 TESAMKKIEETNTLVFLCDPRASKRRIKEAVKETYGVVAQRVNTLIRVDGKKKAYV 147
TE AMKK + NTLVF+ D +A+K + K+AV++ Y + NTLIR DG+KK +V
Sbjct 148 TELAMKKKDNNNTLVFIVDVKANKYQTKQAVEKFYDIDMTSANTLIRPDGEKKTFV 203
> Hs22045978
Length=692
Score = 53.1 bits (126), Expect = 3e-07, Method: Composition-based stats.
Identities = 36/104 (34%), Positives = 56/104 (53%), Gaps = 17/104 (16%)
Query 36 RTATAKARYTVRFRRPRTLKKPRTPKAPRLARLWAANPIAALQRNDKYRVLQHPVTTESA 95
+ A K+R+ R+PR P+T + R R D +++ P TTESA
Sbjct 73 KGAEEKSRHCTSRRQPRY---PQTSTSRR-------------SRPDHCAIIKFPQTTESA 116
Query 96 MKKIEETNTLVFLCDPRASKRRIKEAVKETYGVVAQRVNTLIRV 139
MKK E NTLV++ D +A+KR+IK+A ++ +V+T IR+
Sbjct 117 MKK-REDNTLVYIVDVKANKRQIKQAARKLTNTSVVKVHTPIRL 159
> Hs17450912
Length=140
Score = 51.6 bits (122), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 24/48 (50%), Positives = 35/48 (72%), Gaps = 0/48 (0%)
Query 81 DKYRVLQHPVTTESAMKKIEETNTLVFLCDPRASKRRIKEAVKETYGV 128
D +++ P+TTESAMKKIE++NTLV D +A K +IK+AVK+ +
Sbjct 43 DHCAIIKFPLTTESAMKKIEDSNTLVITVDVKAKKHQIKQAVKKLCDI 90
> Hs22063285
Length=132
Score = 48.9 bits (115), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 31/75 (41%), Positives = 43/75 (57%), Gaps = 3/75 (4%)
Query 91 TTESAMKKIEETNTLVFLCDPRASKRRIKEAVKETYGVVAQRVNTLIRVDGKKKAYVRLT 150
T ES MK E+ + VF D +A+K +IK+AVK+ VN LIR +++
Sbjct 61 TAESTMK-TEDNHMPVFTVDAKANKCQIKQAVKKLCDTDVANVNNLIRAWWREEGIHSTV 119
Query 151 PEFDALDVANKIGII 165
+ ALD+ANKIGII
Sbjct 120 --YHALDIANKIGII 132
> Hs22042184
Length=365
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 22/46 (47%), Positives = 32/46 (69%), Gaps = 2/46 (4%)
Query 78 QRNDKYRVLQH--PVTTESAMKKIEETNTLVFLCDPRASKRRIKEA 121
Q + + QH P+TTESAMKK E+ NTLVF+ D +AS+ ++K+
Sbjct 22 QEKQPWLLCQHQVPLTTESAMKKKEDNNTLVFIVDVKASEHQVKQG 67
> Hs22043659
Length=112
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 19/43 (44%), Positives = 30/43 (69%), Gaps = 2/43 (4%)
Query 93 ESAMKKIEETNTLVFLCDPRASKRRIKEAVKETYGVVAQRVNT 135
E+AMK E NTLVF D +A+K ++++A+KE + + +VNT
Sbjct 69 EAAMK--TEGNTLVFTVDVKANKHQMEQAMKEFHDICVAKVNT 109
> Hs17458937
Length=110
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 19/41 (46%), Positives = 28/41 (68%), Gaps = 0/41 (0%)
Query 81 DKYRVLQHPVTTESAMKKIEETNTLVFLCDPRASKRRIKEA 121
D + P+TTESA KK+E+ NTLVF + +K++IK+A
Sbjct 70 DHCAISTFPLTTESATKKVEDNNTLVFAVVVKPNKQQIKQA 110
> Hs20538039
Length=724
Score = 37.4 bits (85), Expect = 0.014, Method: Composition-based stats.
Identities = 19/35 (54%), Positives = 26/35 (74%), Gaps = 0/35 (0%)
Query 89 PVTTESAMKKIEETNTLVFLCDPRASKRRIKEAVK 123
P+TTE +KK E+ +TLVF+ D + +K RIK AVK
Sbjct 367 PLTTEFMVKKREDNSTLVFIVDVKTNKHRIKLAVK 401
> Hs17474260
Length=110
Score = 34.3 bits (77), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 16/26 (61%), Positives = 19/26 (73%), Gaps = 0/26 (0%)
Query 122 VKETYGVVAQRVNTLIRVDGKKKAYV 147
VK+ Y +VNTLIR DG+KKAYV
Sbjct 85 VKKLYDTDVAKVNTLIRPDGEKKAYV 110
> At4g39880
Length=178
Score = 32.0 bits (71), Expect = 0.50, Method: Compositional matrix adjust.
Identities = 17/34 (50%), Positives = 22/34 (64%), Gaps = 1/34 (2%)
Query 111 PRASKRRIKEAVKETYGVVAQRVNTLIRVDGKKK 144
P ASK IK ++ YG ++VNTL +DGKKK
Sbjct 37 PSASKIEIKRVLESLYGFDVEKVNTL-NMDGKKK 69
> Hs18551250
Length=111
Score = 30.8 bits (68), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 19/61 (31%), Positives = 32/61 (52%), Gaps = 7/61 (11%)
Query 48 FRRPRTLKKPRTPKAPRLARLWAANPIAALQRNDKYRVLQHPVTTESAMKKIEETNTLVF 107
F+RP+TL+ R P+ ++ I + +++ P+TTE A KK ++ NTL
Sbjct 45 FQRPKTLRLWRRPR-------YSQKSIPGRNKLGHCVIIKFPLTTELATKKTDDNNTLCS 97
Query 108 L 108
L
Sbjct 98 L 98
> Hs18605389
Length=117
Score = 29.3 bits (64), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 15/40 (37%), Positives = 23/40 (57%), Gaps = 0/40 (0%)
Query 83 YRVLQHPVTTESAMKKIEETNTLVFLCDPRASKRRIKEAV 122
+ L ++ E AMKK E+ NTL D +A+K + K+ V
Sbjct 69 FITLPDSLSIECAMKKTEDNNTLEVTVDGKANKHQSKQDV 108
> CE29543
Length=445
Score = 28.9 bits (63), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 26/49 (53%), Gaps = 0/49 (0%)
Query 57 PRTPKAPRLARLWAANPIAALQRNDKYRVLQHPVTTESAMKKIEETNTL 105
PRTP++ RLA + AL + + Q P+ +S ++K + NTL
Sbjct 361 PRTPRSSRLAEVDEMGRYHALYDDSHSQTPQKPMRIKSTLRKYLDKNTL 409
> CE27608
Length=724
Score = 28.5 bits (62), Expect = 7.1, Method: Composition-based stats.
Identities = 19/68 (27%), Positives = 30/68 (44%), Gaps = 8/68 (11%)
Query 98 KIEETNTLVFLCDPRASKRRIKEAVKETYGVVAQRVNTLIRVDGKKKAYVRLTPEFDALD 157
+I E L FL + ++ V E G + + +R+DG + L+P D
Sbjct 621 RISEIEELAFL--------KSRDQVTELIGQLVEEQRITVRIDGDMVFWTELSPVPSKND 672
Query 158 VANKIGII 165
V NKI I+
Sbjct 673 VENKIRIV 680
> CE13433
Length=159
Score = 28.1 bits (61), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 22/78 (28%), Positives = 39/78 (50%), Gaps = 6/78 (7%)
Query 64 RLARLWA-ANPIAALQRNDKYR-VLQHPVTTESAMKKIEETNTLVFLCDPRASKRRIKEA 121
RLARLW NP + D + V++ P + + + N + F DPR S+ I+E
Sbjct 4 RLARLWQPGNPQRRVFLPDFWMAVVESPSVGRNRLPR----NCVKFEVDPRMSRHDIREY 59
Query 122 VKETYGVVAQRVNTLIRV 139
+ + Y + + V T +++
Sbjct 60 LTKIYDLPVRDVRTEVQM 77
Lambda K H
0.316 0.129 0.361
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2353551590
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40