bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1014_orf1
Length=88
Score E
Sequences producing significant alignments: (Bits) Value
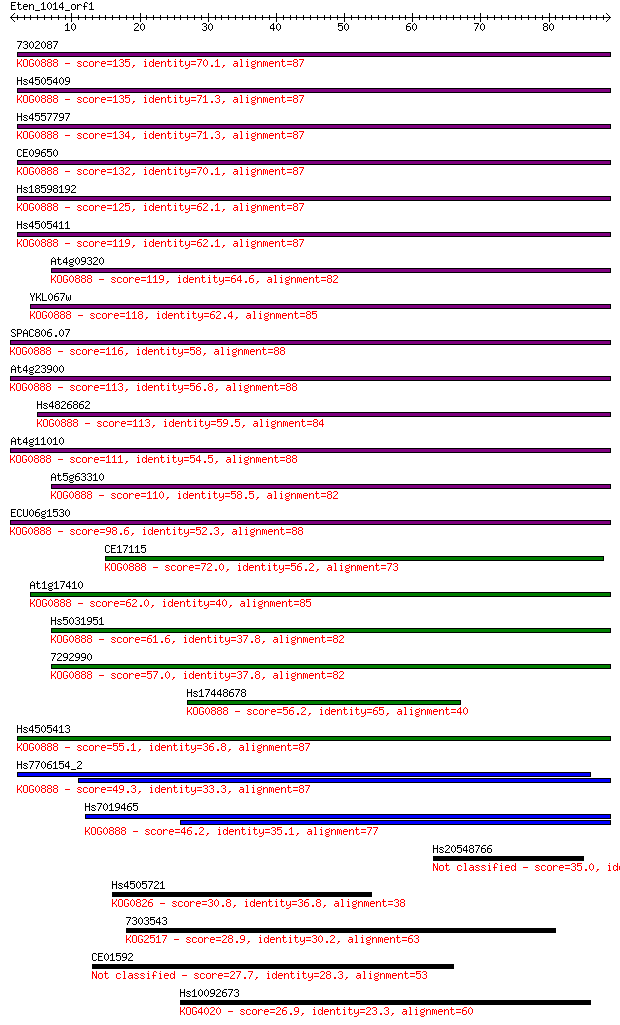
7302087 135 2e-32
Hs4505409 135 2e-32
Hs4557797 134 3e-32
CE09650 132 1e-31
Hs18598192 125 3e-29
Hs4505411 119 1e-27
At4g09320 119 2e-27
YKL067w 118 3e-27
SPAC806.07 116 1e-26
At4g23900 113 8e-26
Hs4826862 113 1e-25
At4g11010 111 3e-25
At5g63310 110 6e-25
ECU06g1530 98.6 2e-21
CE17115 72.0 2e-13
At1g17410 62.0 3e-10
Hs5031951 61.6 3e-10
7292990 57.0 7e-09
Hs17448678 56.2 1e-08
Hs4505413 55.1 3e-08
Hs7706154_2 49.3 2e-06
Hs7019465 46.2 1e-05
Hs20548766 35.0 0.031
Hs4505721 30.8 0.67
7303543 28.9 2.4
CE01592 27.7 6.0
Hs10092673 26.9 9.8
> 7302087
Length=172
Score = 135 bits (340), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 61/87 (70%), Positives = 72/87 (82%), Gaps = 0/87 (0%)
Query 2 SSQTLEQHYRDLSDKPFFRVLMNYMSSGPVVAMVWEGTDVVKQARKMLGETRPLDSQPGT 61
S + LE+HY DLS +PFF L+NYM+SGPVV MVWEG +VVK R+MLG T P DS PGT
Sbjct 64 SKELLEKHYADLSARPFFPGLVNYMNSGPVVPMVWEGLNVVKTGRQMLGATNPADSLPGT 123
Query 62 IRGDFCIDVGRNLIHGSDSVEAAEREI 88
IRGDFCI VGRN+IHGSD+VE+AE+EI
Sbjct 124 IRGDFCIQVGRNIIHGSDAVESAEKEI 150
> Hs4505409
Length=152
Score = 135 bits (340), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 62/87 (71%), Positives = 73/87 (83%), Gaps = 0/87 (0%)
Query 2 SSQTLEQHYRDLSDKPFFRVLMNYMSSGPVVAMVWEGTDVVKQARKMLGETRPLDSQPGT 61
S + L+QHY DL D+PFF L+ YM+SGPVVAMVWEG +VVK R MLGET P DS+PGT
Sbjct 44 SEEHLKQHYIDLKDRPFFPGLVKYMNSGPVVAMVWEGLNVVKTGRVMLGETNPADSKPGT 103
Query 62 IRGDFCIDVGRNLIHGSDSVEAAEREI 88
IRGDFCI VGRN+IHGSDSV++AE+EI
Sbjct 104 IRGDFCIQVGRNIIHGSDSVKSAEKEI 130
> Hs4557797
Length=152
Score = 134 bits (338), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 62/87 (71%), Positives = 71/87 (81%), Gaps = 0/87 (0%)
Query 2 SSQTLEQHYRDLSDKPFFRVLMNYMSSGPVVAMVWEGTDVVKQARKMLGETRPLDSQPGT 61
S L++HY DL D+PFF L+ YM SGPVVAMVWEG +VVK R MLGET P DS+PGT
Sbjct 44 SEDLLKEHYVDLKDRPFFAGLVKYMHSGPVVAMVWEGLNVVKTGRVMLGETNPADSKPGT 103
Query 62 IRGDFCIDVGRNLIHGSDSVEAAEREI 88
IRGDFCI VGRN+IHGSDSVE+AE+EI
Sbjct 104 IRGDFCIQVGRNIIHGSDSVESAEKEI 130
> CE09650
Length=153
Score = 132 bits (333), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 61/87 (70%), Positives = 68/87 (78%), Gaps = 0/87 (0%)
Query 2 SSQTLEQHYRDLSDKPFFRVLMNYMSSGPVVAMVWEGTDVVKQARKMLGETRPLDSQPGT 61
S LE HY+DL DKPFF L+ YMSSGPVVAMVW+G DVVKQ R MLG T PL S PGT
Sbjct 44 SKAHLEVHYQDLKDKPFFPSLIEYMSSGPVVAMVWQGLDVVKQGRSMLGATNPLASAPGT 103
Query 62 IRGDFCIDVGRNLIHGSDSVEAAEREI 88
IRGDFCI GRN+ HGSD+V++A REI
Sbjct 104 IRGDFCIQTGRNICHGSDAVDSANREI 130
> Hs18598192
Length=169
Score = 125 bits (313), Expect = 3e-29, Method: Compositional matrix adjust.
Identities = 54/87 (62%), Positives = 71/87 (81%), Gaps = 0/87 (0%)
Query 2 SSQTLEQHYRDLSDKPFFRVLMNYMSSGPVVAMVWEGTDVVKQARKMLGETRPLDSQPGT 61
S + L +HY +L ++PF+ L+ YM+SGPVVAMVW+G DVV+ +R ++G T P D+ PGT
Sbjct 61 SEELLREHYAELRERPFYGRLVKYMASGPVVAMVWQGLDVVRTSRALIGATNPADAPPGT 120
Query 62 IRGDFCIDVGRNLIHGSDSVEAAEREI 88
IRGDFCI+VG+NLIHGSDSVE+A REI
Sbjct 121 IRGDFCIEVGKNLIHGSDSVESARREI 147
> Hs4505411
Length=168
Score = 119 bits (298), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 54/87 (62%), Positives = 70/87 (80%), Gaps = 1/87 (1%)
Query 2 SSQTLEQHYRDLSDKPFFRVLMNYMSSGPVVAMVWEGTDVVKQARKMLGETRPLDSQPGT 61
S + L +HY +L ++PF+ L+ YM+SGPVVAMVW+G DVV+ +R ++G T P D+ PGT
Sbjct 61 SEELLREHYAELRERPFYGRLVKYMASGPVVAMVWQGLDVVRTSRALIGATNPADAPPGT 120
Query 62 IRGDFCIDVGRNLIHGSDSVEAAEREI 88
IRGDFCI+VG NLIHGSDSVE+A REI
Sbjct 121 IRGDFCIEVG-NLIHGSDSVESARREI 146
> At4g09320
Length=149
Score = 119 bits (297), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 53/82 (64%), Positives = 66/82 (80%), Gaps = 0/82 (0%)
Query 7 EQHYRDLSDKPFFRVLMNYMSSGPVVAMVWEGTDVVKQARKMLGETRPLDSQPGTIRGDF 66
E+HY DLS K FF L++Y+ SGPVVAM+WEG +VV RK++G T P S+PGTIRGDF
Sbjct 46 EKHYEDLSSKSFFSGLVDYIVSGPVVAMIWEGKNVVLTGRKIIGATNPAASEPGTIRGDF 105
Query 67 CIDVGRNLIHGSDSVEAAEREI 88
ID+GRN+IHGSDSVE+A +EI
Sbjct 106 AIDIGRNVIHGSDSVESARKEI 127
> YKL067w
Length=153
Score = 118 bits (295), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 53/85 (62%), Positives = 67/85 (78%), Gaps = 0/85 (0%)
Query 4 QTLEQHYRDLSDKPFFRVLMNYMSSGPVVAMVWEGTDVVKQARKMLGETRPLDSQPGTIR 63
+ LEQHY + KPFF ++++M SGP++A VWEG DVV+Q R +LG T PL S PGTIR
Sbjct 47 KLLEQHYAEHVGKPFFPKMVSFMKSGPILATVWEGKDVVRQGRTILGATNPLGSAPGTIR 106
Query 64 GDFCIDVGRNLIHGSDSVEAAEREI 88
GDF ID+GRN+ HGSDSV++AEREI
Sbjct 107 GDFGIDLGRNVCHGSDSVDSAEREI 131
> SPAC806.07
Length=151
Score = 116 bits (290), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 51/88 (57%), Positives = 65/88 (73%), Gaps = 0/88 (0%)
Query 1 PSSQTLEQHYRDLSDKPFFRVLMNYMSSGPVVAMVWEGTDVVKQARKMLGETRPLDSQPG 60
PS +E+HY + KPF+ L+ +M+SGPV AM+WEG VK R MLG + PLDS PG
Sbjct 42 PSRDLVEEHYAEHKGKPFYEKLVGFMASGPVCAMIWEGKQAVKTGRLMLGASNPLDSAPG 101
Query 61 TIRGDFCIDVGRNLIHGSDSVEAAEREI 88
TIRGD+ ID+GRN+ HGSDS+E+A REI
Sbjct 102 TIRGDYGIDLGRNVCHGSDSIESANREI 129
> At4g23900
Length=237
Score = 113 bits (283), Expect = 8e-26, Method: Compositional matrix adjust.
Identities = 50/88 (56%), Positives = 64/88 (72%), Gaps = 0/88 (0%)
Query 1 PSSQTLEQHYRDLSDKPFFRVLMNYMSSGPVVAMVWEGTDVVKQARKMLGETRPLDSQPG 60
PS ++HY DL ++PFF L N++SSGPVVAMVWEG V++ RK++G T P S+PG
Sbjct 126 PSKGFAQKHYHDLKERPFFNGLCNFLSSGPVVAMVWEGEGVIRYGRKLIGATDPQKSEPG 185
Query 61 TIRGDFCIDVGRNLIHGSDSVEAAEREI 88
TIRGD + VGRN+IHGSD E A+ EI
Sbjct 186 TIRGDLAVVVGRNIIHGSDGPETAKDEI 213
> Hs4826862
Length=187
Score = 113 bits (282), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 50/84 (59%), Positives = 64/84 (76%), Gaps = 0/84 (0%)
Query 5 TLEQHYRDLSDKPFFRVLMNYMSSGPVVAMVWEGTDVVKQARKMLGETRPLDSQPGTIRG 64
L +HY+DL KPF+ L+ YMSSGPVVAMVWEG +VV+ +R M+G T ++ PGTIRG
Sbjct 80 VLAEHYQDLRRKPFYPALIRYMSSGPVVAMVWEGYNVVRASRAMIGHTDSAEAAPGTIRG 139
Query 65 DFCIDVGRNLIHGSDSVEAAEREI 88
DF + + RN+IH SDSVE A+REI
Sbjct 140 DFSVHISRNVIHASDSVEGAQREI 163
> At4g11010
Length=238
Score = 111 bits (278), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 48/88 (54%), Positives = 64/88 (72%), Gaps = 0/88 (0%)
Query 1 PSSQTLEQHYRDLSDKPFFRVLMNYMSSGPVVAMVWEGTDVVKQARKMLGETRPLDSQPG 60
PS ++HY DL ++PFF L +++SSGPV+AMVWEG V++ RK++G T P S+PG
Sbjct 127 PSKDFAQKHYHDLKERPFFNGLCDFLSSGPVIAMVWEGDGVIRYGRKLIGATDPQKSEPG 186
Query 61 TIRGDFCIDVGRNLIHGSDSVEAAEREI 88
TIRGD + VGRN+IHGSD E A+ EI
Sbjct 187 TIRGDLAVTVGRNIIHGSDGPETAKDEI 214
> At5g63310
Length=231
Score = 110 bits (275), Expect = 6e-25, Method: Compositional matrix adjust.
Identities = 48/82 (58%), Positives = 62/82 (75%), Gaps = 0/82 (0%)
Query 7 EQHYRDLSDKPFFRVLMNYMSSGPVVAMVWEGTDVVKQARKMLGETRPLDSQPGTIRGDF 66
E+HY+DLS K FF L+ Y++SGPVV M WEG VV ARK++G+T PL ++PGTIRGD
Sbjct 128 EEHYKDLSAKSFFPNLIEYITSGPVVCMAWEGVGVVASARKLIGKTDPLQAEPGTIRGDL 187
Query 67 CIDVGRNLIHGSDSVEAAEREI 88
+ GRN++HGSDS E +REI
Sbjct 188 AVQTGRNIVHGSDSPENGKREI 209
> ECU06g1530
Length=147
Score = 98.6 bits (244), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 46/88 (52%), Positives = 58/88 (65%), Gaps = 0/88 (0%)
Query 1 PSSQTLEQHYRDLSDKPFFRVLMNYMSSGPVVAMVWEGTDVVKQARKMLGETRPLDSQPG 60
P + LE HY LS PFF ++ M SG V+AMVW G D V RK++GET P + G
Sbjct 40 PKREVLETHYSHLSSMPFFSEMVEDMMSGMVLAMVWVGKDAVSIGRKLIGETNPQAASVG 99
Query 61 TIRGDFCIDVGRNLIHGSDSVEAAEREI 88
TIRGD+ + G+N+IHGSD VE AE+EI
Sbjct 100 TIRGDYGVSTGKNIIHGSDCVENAEKEI 127
> CE17115
Length=118
Score = 72.0 bits (175), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 41/76 (53%), Positives = 51/76 (67%), Gaps = 7/76 (9%)
Query 15 DKPFFRVLMNYMSSGPVVAMVWEGTDVVKQARKMLGETRPLDSQPGTIRGDFCIDVGRNL 74
DKPFF +L++YMSSGPVVAM+WEG DVVK+AR +LGE + + G R F V R+
Sbjct 32 DKPFFPLLIDYMSSGPVVAMLWEGCDVVKRARVILGE----ELEVGEFRSIFYDLVVRDT 87
Query 75 ---IHGSDSVEAAERE 87
H SDSV +A RE
Sbjct 88 HKGCHCSDSVASANRE 103
> At1g17410
Length=162
Score = 62.0 bits (149), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 34/88 (38%), Positives = 49/88 (55%), Gaps = 3/88 (3%)
Query 4 QTLEQHYRDLSDKPFFRVLMNYMSSGPVVAMVWEGTDVVKQARKMLGET---RPLDSQPG 60
+T Y + S + FF L+ YM+SGPV+ MV E + V R ++G T + S P
Sbjct 37 ETASAFYEEHSSRSFFPHLVTYMTSGPVLVMVLEKRNAVSDWRDLIGPTDAEKAKISHPH 96
Query 61 TIRGDFCIDVGRNLIHGSDSVEAAEREI 88
+IR + +N +HGSDS +AEREI
Sbjct 97 SIRALCGKNSQKNCVHGSDSTSSAEREI 124
> Hs5031951
Length=194
Score = 61.6 bits (148), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 31/85 (36%), Positives = 51/85 (60%), Gaps = 3/85 (3%)
Query 7 EQHYRDLSDKPFFRVLMNYMSSGPVVAMVWEGTDVVKQARKMLGETRPLDSQ---PGTIR 63
++ YR+ + F++ L+ +M+SGP+ A + D ++ R ++G TR ++ P +IR
Sbjct 65 QRFYREHEGRFFYQRLVEFMASGPIRAYILAHKDAIQLWRTLMGPTRVFRARHVAPDSIR 124
Query 64 GDFCIDVGRNLIHGSDSVEAAEREI 88
G F + RN HGSDSV +A REI
Sbjct 125 GSFGLTDTRNTTHGSDSVVSASREI 149
> 7292990
Length=127
Score = 57.0 bits (136), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 31/85 (36%), Positives = 47/85 (55%), Gaps = 3/85 (3%)
Query 7 EQHYRDLSDKPFFRVLMNYMSSGPVVAMVWEGTDVVKQARKMLGET---RPLDSQPGTIR 63
E+ Y + K F+ L ++M+SGP A++ + +++ R +LG T R + S P IR
Sbjct 22 ERFYAEHKGKFFYHRLTSFMNSGPSYALILQSETCIQKWRSLLGPTKVFRAVYSDPNCIR 81
Query 64 GDFCIDVGRNLIHGSDSVEAAEREI 88
+ I RN HGSDS +A REI
Sbjct 82 ALYGISDTRNACHGSDSEASALREI 106
> Hs17448678
Length=132
Score = 56.2 bits (134), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 26/40 (65%), Positives = 31/40 (77%), Gaps = 0/40 (0%)
Query 27 SSGPVVAMVWEGTDVVKQARKMLGETRPLDSQPGTIRGDF 66
+SGPV+AMVWEG + VK R MLGET P+DS+P TI DF
Sbjct 73 NSGPVLAMVWEGLNAVKTGRVMLGETNPVDSKPHTICRDF 112
> Hs4505413
Length=212
Score = 55.1 bits (131), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 32/90 (35%), Positives = 46/90 (51%), Gaps = 3/90 (3%)
Query 2 SSQTLEQHYRDLSDKPFFRVLMNYMSSGPVVAMVWEGTDVVKQARKMLGETRPL---DSQ 58
S + Y + K FF L YMSSGP+VAM+ + ++LG L ++
Sbjct 50 SPEQCSNFYVEKYGKMFFPNLTAYMSSGPLVAMILARHKAISYWLELLGPNNSLVAKETH 109
Query 59 PGTIRGDFCIDVGRNLIHGSDSVEAAEREI 88
P ++R + D RN +HGS+ AAEREI
Sbjct 110 PDSLRAIYGTDDLRNALHGSNDFAAAEREI 139
> Hs7706154_2
Length=373
Score = 49.3 bits (116), Expect = 2e-06, Method: Composition-based stats.
Identities = 26/88 (29%), Positives = 52/88 (59%), Gaps = 4/88 (4%)
Query 2 SSQTLEQHYRDLSDKPFFRVLMNYMSSGPVVAMVWEGTDVVKQARKMLGETRPLDSQ--- 58
+ + +E+ Y ++ K F++ L+ +S GP + M+ + V + R+++G T P +++
Sbjct 274 TPEQIEKIYPKVTGKDFYKDLLEMLSVGPSMVMILTKWNAVAEWRRLMGPTDPEEAKLLS 333
Query 59 PGTIRGDFCIDVGRNLIHG-SDSVEAAE 85
P +IR F I +N++HG S++ EA E
Sbjct 334 PDSIRAQFGISKLKNIVHGASNAYEAKE 361
Score = 39.3 bits (90), Expect = 0.002, Method: Composition-based stats.
Identities = 24/82 (29%), Positives = 48/82 (58%), Gaps = 4/82 (4%)
Query 11 RDLSDKPFFRVLMNYMSSGPVVAMVWEGTDVVKQARKMLGE---TRPLDSQPGTIRGDFC 67
++ ++ +F L+ M+SGP +A+V + ++ +++LG ++ P ++ F
Sbjct 148 KEYENEDYFNKLIENMTSGPSLALVLLRDNGLQYWKQLLGPRTVEEAIEYFPESLCAQFA 207
Query 68 ID-VGRNLIHGSDSVEAAEREI 88
+D + N ++GSDS+E AEREI
Sbjct 208 MDSLPVNQLYGSDSLETAEREI 229
> Hs7019465
Length=376
Score = 46.2 bits (108), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 27/83 (32%), Positives = 44/83 (53%), Gaps = 9/83 (10%)
Query 12 DLSDKPFFRVLMNYMSSGPVVAMVWEGTDVVKQARKMLGETRPLDSQPG------TIRGD 65
D +PFF L+ ++++GP++AM D + + +++LG P +S +IR
Sbjct 139 DHQSRPFFNELIQFITTGPIIAMEILRDDAICEWKRLLG---PANSGVARTDASESIRAL 195
Query 66 FCIDVGRNLIHGSDSVEAAEREI 88
F D RN HG DS +A RE+
Sbjct 196 FGTDGIRNAAHGPDSFASAAREM 218
Score = 32.3 bits (72), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 21/66 (31%), Positives = 31/66 (46%), Gaps = 3/66 (4%)
Query 26 MSSGPVVAMVWEGTDVVKQARKMLGETRPLDS---QPGTIRGDFCIDVGRNLIHGSDSVE 82
M SGP VAM + + K R+ G P + +PGT+R F +N +H +D E
Sbjct 302 MYSGPCVAMEIQQNNATKTFREFCGPADPEIARHLRPGTLRAIFGKTKIQNAVHCTDLPE 361
Query 83 AAEREI 88
E+
Sbjct 362 DGLLEV 367
> Hs20548766
Length=705
Score = 35.0 bits (79), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 15/22 (68%), Positives = 18/22 (81%), Gaps = 0/22 (0%)
Query 63 RGDFCIDVGRNLIHGSDSVEAA 84
R FCI+ GRN+IHGS SVE+A
Sbjct 646 RKSFCIEDGRNIIHGSMSVESA 667
> Hs4505721
Length=359
Score = 30.8 bits (68), Expect = 0.67, Method: Composition-based stats.
Identities = 14/38 (36%), Positives = 24/38 (63%), Gaps = 3/38 (7%)
Query 16 KPFFRVLMNYMSSGPVVAMVWEGTDVVKQARKMLGETR 53
K F+R ++++ P V M WEG +V+Q R +LG+ +
Sbjct 153 KRFYRA---FLAAYPFVNMAWEGWFLVQQLRYILGKAQ 187
> 7303543
Length=596
Score = 28.9 bits (63), Expect = 2.4, Method: Composition-based stats.
Identities = 19/67 (28%), Positives = 30/67 (44%), Gaps = 10/67 (14%)
Query 18 FFRVLMNYM----SSGPVVAMVWEGTDVVKQARKMLGETRPLDSQPGTIRGDFCIDVGRN 73
FFR+ +N + S+ + V EG M+GE QP ++ G C+ G+N
Sbjct 222 FFRLPLNILPRIRSNSEIFGYVLEGPLHGTPIAAMMGE------QPASLLGQLCVKAGQN 275
Query 74 LIHGSDS 80
+ DS
Sbjct 276 VCTLDDS 282
> CE01592
Length=233
Score = 27.7 bits (60), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 15/53 (28%), Positives = 24/53 (45%), Gaps = 0/53 (0%)
Query 13 LSDKPFFRVLMNYMSSGPVVAMVWEGTDVVKQARKMLGETRPLDSQPGTIRGD 65
S P + + SS + + +G D+VKQA + G R D+Q + D
Sbjct 44 FSTPPIYIFKVFTRSSFRLPCRILDGMDIVKQATQAPGLNRQFDTQTNVLAAD 96
> Hs10092673
Length=367
Score = 26.9 bits (58), Expect = 9.8, Method: Composition-based stats.
Identities = 14/60 (23%), Positives = 33/60 (55%), Gaps = 0/60 (0%)
Query 26 MSSGPVVAMVWEGTDVVKQARKMLGETRPLDSQPGTIRGDFCIDVGRNLIHGSDSVEAAE 85
+S+G VA+VW+ + V+ + ++ + + L G + + + + L+ GS ++ +AE
Sbjct 6 ISAGQFVAVVWDKSSPVEALKGLVDKLQALTGNEGRVSVENIKQLLQCLVPGSTTLHSAE 65
Lambda K H
0.318 0.135 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1184307974
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40