bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1037_orf1
Length=116
Score E
Sequences producing significant alignments: (Bits) Value
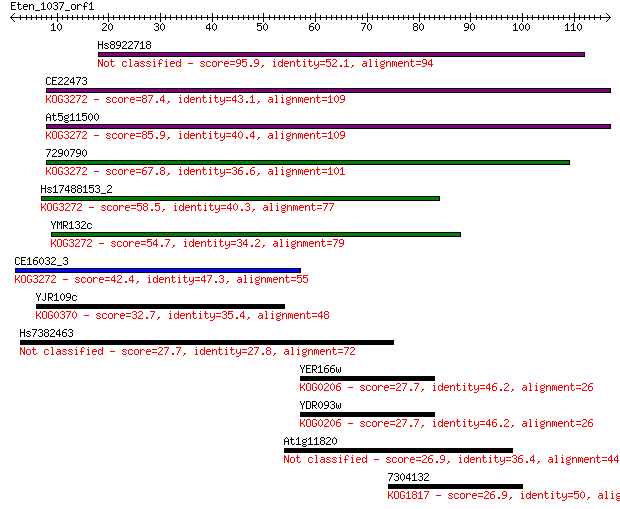
Hs8922718 95.9 2e-20
CE22473 87.4 6e-18
At5g11500 85.9 2e-17
7290790 67.8 4e-12
Hs17488153_2 58.5 3e-09
YMR132c 54.7 4e-08
CE16032_3 42.4 2e-04
YJR109c 32.7 0.16
Hs7382463 27.7 5.5
YER166w 27.7 5.8
YDR093w 27.7 6.0
At1g11820 26.9 9.6
7304132 26.9 9.7
> Hs8922718
Length=140
Score = 95.9 bits (237), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 49/94 (52%), Positives = 64/94 (68%), Gaps = 0/94 (0%)
Query 18 EACQLTKQNSIEGCKQSEVKIVYTPAGNLKKTANMETGQVGFKQQSLVKEVQGVKKEKEL 77
+ L K NSI+GCK + V +VYTP NLKKTA+M+ GQ+GF +Q VK V KK E+
Sbjct 2 DCAHLVKANSIQGCKMNNVNVVYTPWSNLKKTADMDVGQIGFHRQKDVKIVTVEKKVNEI 61
Query 78 LKALEKTRAEPAVDLAANRLERDRRERSERKARL 111
L LEKT+ E DLAA + RDR ER+E+KA++
Sbjct 62 LNRLEKTKVERFPDLAAEKECRDREERNEKKAQI 95
> CE22473
Length=210
Score = 87.4 bits (215), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 47/109 (43%), Positives = 64/109 (58%), Gaps = 2/109 (1%)
Query 8 FEELPAQVIAEACQLTKQNSIEGCKQSEVKIVYTPAGNLKKTANMETGQVGFKQQSLVKE 67
+ +P ++ + CQL KQNSIEGCK + V IVYT NLKKT +M GQVGF VK
Sbjct 59 IDSIPEALLIDCCQLVKQNSIEGCKLNNVAIVYTMWSNLKKTGDMAVGQVGFHSHKQVKH 118
Query 68 VQGVKKEKELLKALEKTRAEPAVDLAANRLERDRRERSERKARLLQQQQ 116
K E++ LEKTR + +D R RD +ER +K + L+Q++
Sbjct 119 TVVPTKINEIVNRLEKTRRKDDIDYKEERDARDAKER--QKLKKLEQER 165
> At5g11500
Length=215
Score = 85.9 bits (211), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 44/109 (40%), Positives = 68/109 (62%), Gaps = 0/109 (0%)
Query 8 FEELPAQVIAEACQLTKQNSIEGCKQSEVKIVYTPAGNLKKTANMETGQVGFKQQSLVKE 67
F+++ V+ + QL K NSI+G K + V +VYTP NLKKTA+M+ GQVGF +V+
Sbjct 59 FDDISEGVLEDCAQLVKANSIQGNKVNNVDVVYTPWSNLKKTASMDVGQVGFHNSKMVRT 118
Query 68 VQGVKKEKELLKALEKTRAEPAVDLAANRLERDRRERSERKARLLQQQQ 116
++ K+ E++ L KT+ E DL A R + ER+ERK L ++++
Sbjct 119 IRVEKRVNEIVNRLNKTKVERTPDLRAEREAVNAAERAERKQHLREKKK 167
> 7290790
Length=209
Score = 67.8 bits (164), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 37/102 (36%), Positives = 60/102 (58%), Gaps = 1/102 (0%)
Query 8 FEELPAQVIAEACQLTKQNSIEGCKQSEVKIVYTPAGNLKKTANMETGQVGFKQQSLVKE 67
+++P V+ +A QL K NSI+G K + +++VYT NLKKT +ME GQV + + V+
Sbjct 59 IDDIPTDVLVDAAQLCKANSIQGNKVNNLEVVYTMWENLKKTPDMEPGQVAYHNEKAVRR 118
Query 68 VQGVKKEKELLKALEKTRA-EPAVDLAANRLERDRRERSERK 108
++ K+ E++ L +T+ E D R RD ER ++K
Sbjct 119 IRLEKRINEIVNRLNRTKTEEEHPDFRGLREARDAAERQDQK 160
> Hs17488153_2
Length=238
Score = 58.5 bits (140), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 31/77 (40%), Positives = 45/77 (58%), Gaps = 1/77 (1%)
Query 7 DFEELPAQVIAEACQLTKQNSIEGCKQSEVKIVYTPAGNLKKTANMETGQVGFKQQSLVK 66
+ E++P +V+ + L K NS +GCK + V +V TP NLKKTA+++ Q+GF +Q VK
Sbjct 3 NIEDIPKEVLTDCAHLVKANSTQGCKNN-VNVVNTPRSNLKKTADVDVRQIGFHRQKDVK 61
Query 67 EVQGVKKEKELLKALEK 83
V KK L K
Sbjct 62 IVTMEKKRGFLAHIWNK 78
> YMR132c
Length=208
Score = 54.7 bits (130), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 27/79 (34%), Positives = 45/79 (56%), Gaps = 0/79 (0%)
Query 9 EELPAQVIAEACQLTKQNSIEGCKQSEVKIVYTPAGNLKKTANMETGQVGFKQQSLVKEV 68
+++P +VI + QL K SI+G K + I+ TP NL+K M G+V FK +++
Sbjct 65 DDIPQEVICDCLQLCKSESIQGNKMPQCTILITPWHNLRKNRYMNPGEVSFKSLRQCRKM 124
Query 69 QGVKKEKELLKALEKTRAE 87
+ ++ ++L L KTR E
Sbjct 125 ECGARDNKILNRLAKTRVE 143
> CE16032_3
Length=148
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 26/70 (37%), Positives = 34/70 (48%), Gaps = 15/70 (21%)
Query 2 VFGFSDFEELPAQVIA---------------EACQLTKQNSIEGCKQSEVKIVYTPAGNL 46
VFGF P +I E CQL K+NSI+G K +V++ YT NL
Sbjct 2 VFGFISSTVTPPALIFMGEHQVENEKLFKLEECCQLVKKNSIQGVKMEKVEVNYTLKENL 61
Query 47 KKTANMETGQ 56
KK M+TG+
Sbjct 62 KKVKGMQTGE 71
> YJR109c
Length=1118
Score = 32.7 bits (73), Expect = 0.16, Method: Composition-based stats.
Identities = 17/48 (35%), Positives = 28/48 (58%), Gaps = 0/48 (0%)
Query 6 SDFEELPAQVIAEACQLTKQNSIEGCKQSEVKIVYTPAGNLKKTANME 53
S+ +EL AQ ++ A Q+ + S++G K+ E ++V GN NME
Sbjct 207 SEMKELAAQSLSLAPQILVEKSLKGWKEVEYEVVRDRVGNCITVCNME 254
> Hs7382463
Length=679
Score = 27.7 bits (60), Expect = 5.5, Method: Composition-based stats.
Identities = 20/75 (26%), Positives = 33/75 (44%), Gaps = 6/75 (8%)
Query 3 FGFSDFEELPAQVIAEACQLTKQNSIEGCKQSEVKIVYTPAGNLKKTANMETGQV---GF 59
F D EE P + + L ++ SI+ V++ P GNL + + + Q+ G
Sbjct 319 FKLGDLEEAPERERLPSVDLKEETSIDSTVNGAVQL---PNGNLVQFSQAVSNQINSSGH 375
Query 60 KQQSLVKEVQGVKKE 74
Q V + G+ KE
Sbjct 376 SQYHTVHKDSGLYKE 390
> YER166w
Length=1571
Score = 27.7 bits (60), Expect = 5.8, Method: Composition-based stats.
Identities = 12/26 (46%), Positives = 19/26 (73%), Gaps = 0/26 (0%)
Query 57 VGFKQQSLVKEVQGVKKEKELLKALE 82
VG ++ L+ E+QG++KE E+L LE
Sbjct 816 VGKTKKGLIIEMQGIQKEFEILNILE 841
> YDR093w
Length=1612
Score = 27.7 bits (60), Expect = 6.0, Method: Composition-based stats.
Identities = 12/26 (46%), Positives = 18/26 (69%), Gaps = 0/26 (0%)
Query 57 VGFKQQSLVKEVQGVKKEKELLKALE 82
VG + L+ E+QGV+KE ++L LE
Sbjct 861 VGSSKSGLIVEIQGVQKEFQVLNVLE 886
> At1g11820
Length=511
Score = 26.9 bits (58), Expect = 9.6, Method: Composition-based stats.
Identities = 16/44 (36%), Positives = 22/44 (50%), Gaps = 0/44 (0%)
Query 54 TGQVGFKQQSLVKEVQGVKKEKELLKALEKTRAEPAVDLAANRL 97
T V F Q V V+ + ELLKAL KT+ + + N+L
Sbjct 59 TELVKFLQAQKVNHVRLYDADPELLKALAKTKVRVIISVPNNQL 102
> 7304132
Length=1327
Score = 26.9 bits (58), Expect = 9.7, Method: Composition-based stats.
Identities = 13/26 (50%), Positives = 18/26 (69%), Gaps = 0/26 (0%)
Query 74 EKELLKALEKTRAEPAVDLAANRLER 99
E+E + LEKTRA+P V+ +RL R
Sbjct 215 ERERNETLEKTRAKPKVETERDRLLR 240
Lambda K H
0.311 0.126 0.331
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1174970866
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40