bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
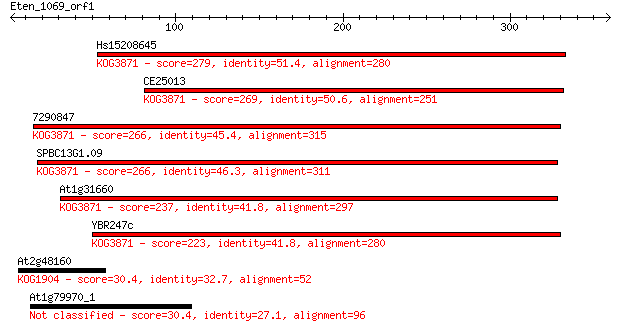
Query= Eten_1069_orf1
Length=359
Score E
Sequences producing significant alignments: (Bits) Value
Hs15208645 279 5e-75
CE25013 269 9e-72
7290847 266 4e-71
SPBC13G1.09 266 6e-71
At1g31660 237 3e-62
YBR247c 223 4e-58
At2g48160 30.4 5.4
At1g79970_1 30.4 6.0
> Hs15208645
Length=301
Score = 279 bits (714), Expect = 5e-75, Method: Compositional matrix adjust.
Identities = 144/284 (50%), Positives = 198/284 (69%), Gaps = 7/284 (2%)
Query 53 NLADLVMEQL-KKQTENKDKSQEER--VESSLSPKVVEVYTAMAPFLARYRSGKMPKAFK 109
LAD++ME+L +KQTE + E L P+V+EVY + L++YRSGK+PKAFK
Sbjct 10 TLADIIMEKLTEKQTEVETVMSEVSGFPMPQLDPRVLEVYRGVREVLSKYRSGKLPKAFK 69
Query 110 IIPRLHAWEEILVLTNPTTWSKQATREATRIFCSNLREAMAQRFLCTVLLPAVRDDIAEN 169
IIP L WE+IL +T P W+ A +ATRIF SNL+E MAQRF VLLP VRDD+AE
Sbjct 70 IIPALSNWEQILYVTEPEAWTAAAMYQATRIFASNLKERMAQRFYNLVLLPRVRDDVAEY 129
Query 170 KKLNYHLYMALKKALFKPGAFFKGIYLPLALDG-CTIREAVIVGSVVTKVSIPVLHGAAA 228
K+LN+HLYMALKKALFKPGA+FKGI +PL G CT+REA+IVGS++TK SIPVLH +AA
Sbjct 130 KRLNFHLYMALKKALFKPGAWFKGILIPLCESGTCTLREAIIVGSIITKCSIPVLHSSAA 189
Query 229 LVRLAAVTPDQWIPSVSHMMTVLIDKKYSLPLQAINECVDHFHRFAPSEVKVTVAWHKCL 288
++++A + ++ + S + +L+DKKY+LP + ++ V HF F + ++ V WH+CL
Sbjct 190 MLKIAEM---EYSGANSIFLRLLLDKKYALPYRVLDALVFHFLGFRTEKRELPVLWHQCL 246
Query 289 LVLIQRYKVNLSASHRMKLKAVLQSHFHAKVGPEIRRELAAQQP 332
L L+QRYK +L+ + L +L+ H ++ PEIRREL + P
Sbjct 247 LTLVQRYKADLATDQKEALLELLRLQPHPQLSPEIRRELQSAVP 290
> CE25013
Length=449
Score = 269 bits (687), Expect = 9e-72, Method: Compositional matrix adjust.
Identities = 127/252 (50%), Positives = 176/252 (69%), Gaps = 4/252 (1%)
Query 81 LSPKVVEVYTAMAPFLARYRSGKMPKAFKIIPRLHAWEEILVLTNPTTWSKQATREATRI 140
+ P+VVE+Y + ++++YRSGK+PKAFKIIP++ WE+IL LT P TW+ A +ATR+
Sbjct 179 MDPEVVEMYEQIGQYMSKYRSGKVPKAFKIIPKMINWEQILFLTKPETWTAAAMYQATRL 238
Query 141 FCSNLREAMAQRFLCTVLLPAVRDDIAENKKLNYHLYMALKKALFKPGAFFKGIYLPLAL 200
F SN+ M QRF VLLP +RDDI E KKLNYHLY AL KA++KP AFFKG+ LPL
Sbjct 239 FASNMNPKMCQRFYTLVLLPRLRDDIDEFKKLNYHLYQALCKAIYKPAAFFKGLILPLLE 298
Query 201 DG-CTIREAVIVGSVVTKVSIPVLHGAAALVRLAAVTPDQWIPSVSHMMTVLIDKKYSLP 259
G CT+REAVI SV+TKV IP+ H AAA++R+A + ++ + S + LIDKKY+LP
Sbjct 299 SGTCTLREAVIFSSVLTKVPIPIFHSAAAMLRIAEM---EYTGANSVFLRALIDKKYALP 355
Query 260 LQAINECVDHFHRFAPSEVKVTVAWHKCLLVLIQRYKVNLSASHRMKLKAVLQSHFHAKV 319
+A++ V+HF R E + V WH+CLL L QRYK +L+A + + +++ H H +
Sbjct 356 YRAVDGVVNHFIRLKTDERDMPVLWHQCLLALCQRYKNDLNAEQKAAIYELIRFHGHYLI 415
Query 320 GPEIRRELAAQQ 331
PEIRREL +++
Sbjct 416 SPEIRRELESKE 427
> 7290847
Length=436
Score = 266 bits (681), Expect = 4e-71, Method: Compositional matrix adjust.
Identities = 143/320 (44%), Positives = 204/320 (63%), Gaps = 10/320 (3%)
Query 15 DEETADKDGFVVLDCEEDEEDLVFEKQRTGSSATRATVNLADLVMEQLK-KQTENKDKSQ 73
DEE + D LD +ED+ FE+ + + + T++L+ ++M++++ K+ + K
Sbjct 90 DEEVNETDLMADLDMDEDDV-AAFERFQQPAQEGKRTLHLSKMIMQKIQEKEADIHTKIS 148
Query 74 EE---RVESSLSPKVVEVYTAMAPFLARYRSGKMPKAFKIIPRLHAWEEILVLTNPTTWS 130
+E ++E + PKV E+Y + L RYRSGK+PKAFKIIP+L WE+IL +T P WS
Sbjct 149 DEGSLKIEE-IDPKVKEMYEGVRDVLKRYRSGKIPKAFKIIPKLRNWEQILFITEPHNWS 207
Query 131 KQATREATRIFCSNLREAMAQRFLCTVLLPAVRDDIAENKKLNYHLYMALKKALFKPGAF 190
A + TRIFCS L +AMAQRF VLLP VRDD+ E KKLN HLY ALK+ALFKP AF
Sbjct 208 AAAMFQGTRIFCSVLSQAMAQRFYNLVLLPRVRDDLCEYKKLNMHLYNALKRALFKPAAF 267
Query 191 FKGIYLPLALDG-CTIREAVIVGSVVTKVSIPVLHGAAALVRLAAVTPDQWIPSVSHMMT 249
KGI LPL G CT+REA+I GSVV + SIPVLH +A L+++ + + + S +
Sbjct 268 MKGIILPLLEGGDCTLREAIIFGSVVARSSIPVLHSSACLLKICEMA---YSGANSIFIR 324
Query 250 VLIDKKYSLPLQAINECVDHFHRFAPSEVKVTVAWHKCLLVLIQRYKVNLSASHRMKLKA 309
+DK+Y+LP + ++ V HF RF + ++ V WH+ LL QRYK ++S+ R L
Sbjct 325 YFLDKRYALPYRVVDAAVFHFLRFENDKRELPVLWHQSLLTFAQRYKNDISSEQRDALLQ 384
Query 310 VLQSHFHAKVGPEIRRELAA 329
+L+ H K+ P++RREL A
Sbjct 385 LLKKKSHFKITPDVRRELQA 404
> SPBC13G1.09
Length=449
Score = 266 bits (679), Expect = 6e-71, Method: Compositional matrix adjust.
Identities = 144/320 (45%), Positives = 199/320 (62%), Gaps = 23/320 (7%)
Query 17 ETADKDGF--VVLDCEEDEEDLVFEKQRTGSSATRATVNLADLVMEQLKKQTENKDKSQ- 73
+ AD+D F + +E DL EK T +L+DL+M+++ + E + + +
Sbjct 128 DDADRDLFDRFLPTVSGEEGDLTEEK----------TTSLSDLIMQKIN-EAEARARGEY 176
Query 74 ------EERVESSLSPKVVEVYTAMAPFLARYRSGKMPKAFKIIPRLHAWEEILVLTNPT 127
EE L PKV+EVY+ + L++YRSGK+PKAFKIIP L WE+IL LT P
Sbjct 177 IPSAEEEENALPPLPPKVIEVYSKVGVLLSKYRSGKIPKAFKIIPTLSNWEDILYLTRPD 236
Query 128 TWSKQATREATRIFCSNLREAMAQRFLCTVLLPAVRDDIAENKKLNYHLYMALKKALFKP 187
W+ A EATRIF SNL+ AQ FL ++L VRDDI ENKKLNYHLYMALKKAL+KP
Sbjct 237 MWTPHACYEATRIFISNLKPVQAQHFLTVIILERVRDDIRENKKLNYHLYMALKKALYKP 296
Query 188 GAFFKGIYLPLALDGCTIREAVIVGSVVTKVSIPVLHGAAALVRLAAVTPDQWIPSVSHM 247
++FKG PL + CT+REA I+GS++ KVS+PVLH AAAL+RL T + S
Sbjct 297 SSWFKGFLFPLVQENCTLREAAIIGSILQKVSVPVLHSAAALLRL---TEFDLSGATSVF 353
Query 248 MTVLIDKKYSLPLQAINECVDHFHRFAPSEVKVTVAWHKCLLVLIQRYKVNLSASHRMKL 307
+ +L+DKKY+LP + ++ V +F R+ E + V H+ +LV QRYK +++ + L
Sbjct 354 IRILLDKKYALPYKVLDSLVFYFMRWKSLERPLAVLEHQSMLVFAQRYKFDITPEQKDAL 413
Query 308 KAVLQSHFHAKVGPEIRREL 327
V++ H +GPEIRREL
Sbjct 414 LEVVRLKGHYSIGPEIRREL 433
> At1g31660
Length=442
Score = 237 bits (604), Expect = 3e-62, Method: Compositional matrix adjust.
Identities = 124/298 (41%), Positives = 185/298 (62%), Gaps = 9/298 (3%)
Query 31 EDEEDLVFEKQRTGSSATRATVNLADLVMEQLKKQTENKDKSQEERVESSLSPKVVEVYT 90
ED+E L FE ++ + T L D+++++LK + + D ++EER + + P + ++Y
Sbjct 118 EDDEKL-FESFLNKNAPPQRT--LTDIIIKKLKDK--DADLAEEERPDPKMDPAITKLYK 172
Query 91 AMAPFLARYRSGKMPKAFKIIPRLHAWEEILVLTNPTTWSKQATREATRIFCSNLREAMA 150
+ F++ Y GK+PKAFK++ + WE++L LT P WS A +ATRIF SNL++
Sbjct 173 GVGKFMSEYTVGKLPKAFKLVTSMEHWEDVLYLTEPEKWSPNALYQATRIFASNLKDRQV 232
Query 151 QRFLCTVLLPAVRDDIAENKKLNYHLYMALKKALFKPGAFFKGIYLPLALDG-CTIREAV 209
QRF VLLP VR+DI ++KKL++ LY ALKK+L+KP AF +GI PL G C +REAV
Sbjct 233 QRFYNYVLLPRVREDIRKHKKLHFALYQALKKSLYKPSAFNQGILFPLCKSGTCNLREAV 292
Query 210 IVGSVVTKVSIPVLHGAAALVRLAAVTPDQWIPSVSHMMTVLIDKKYSLPLQAINECVDH 269
I+GS++ K SIP+LH AL RLA + + + S+ + VL++KKY +P + ++ V H
Sbjct 293 IIGSILEKCSIPMLHSCVALNRLAEM---DYCGTTSYFIKVLLEKKYCMPYRVLDALVAH 349
Query 270 FHRFAPSEVKVTVAWHKCLLVLIQRYKVNLSASHRMKLKAVLQSHFHAKVGPEIRREL 327
F RF + V WH+ LL +QRYK + + L+ +LQ H V PEI REL
Sbjct 350 FMRFVDDIRVMPVIWHQSLLTFVQRYKYEILKEDKEHLQTLLQRQKHHLVTPEILREL 407
> YBR247c
Length=483
Score = 223 bits (569), Expect = 4e-58, Method: Compositional matrix adjust.
Identities = 117/311 (37%), Positives = 175/311 (56%), Gaps = 34/311 (10%)
Query 50 ATVNLADLVMEQLK-KQTENKDKSQEERVES-------------------SLSPKVVEVY 89
+ NLAD +M ++ K+++ +D +E + + +L KV++ Y
Sbjct 155 GSYNLADKIMASIREKESQVEDMQDDEPLANEQNTSRGNISSGLKSGEGVALPEKVIKAY 214
Query 90 TAMAPFLARYRSGKMPKAFKIIPRLHAWEEILVLTNPTTWSKQATREATRIFCSNLREAM 149
T + L + GK+PK FK+IP L W++++ +TNP WS EAT++F SNL
Sbjct 215 TTVGSILKTWTHGKLPKLFKVIPSLRNWQDVIYVTNPEEWSPHVVYEATKLFVSNLTAKE 274
Query 150 AQRFLCTVLLPAVRDDI--AENKKLNYHLYMALKKALFKPGAFFKGIYLPLALDGCTIRE 207
+Q+F+ +LL RD+I +E+ LNYH+Y A+KK+L+KP AFFKG PL GC +RE
Sbjct 275 SQKFINLILLERFRDNIETSEDHSLNYHIYRAVKKSLYKPSAFFKGFLFPLVETGCNVRE 334
Query 208 AVIVGSVVTKVSIPVLHGAAALVRLAAVTPDQWIPSVSHMMTVLIDKKYSLPLQAINECV 267
A I GSV+ KVS+P LH +AAL L + + P + + +L+DKKY+LP Q +++CV
Sbjct 335 ATIAGSVLAKVSVPALHSSAALSYLLRLP---FSPPTTVFIKILLDKKYALPYQTVDDCV 391
Query 268 DHFHRF--------APSEVKV-TVAWHKCLLVLIQRYKVNLSASHRMKLKAVLQSHFHAK 318
+F RF +V V WHK L QRYK +++ R L ++ H
Sbjct 392 YYFMRFRILDDGSNGEDATRVLPVIWHKAFLTFAQRYKNDITQDQRDFLLETVRQRGHKD 451
Query 319 VGPEIRRELAA 329
+GPEIRREL A
Sbjct 452 IGPEIRRELLA 462
> At2g48160
Length=1191
Score = 30.4 bits (67), Expect = 5.4, Method: Composition-based stats.
Identities = 17/52 (32%), Positives = 26/52 (50%), Gaps = 0/52 (0%)
Query 6 ASDAEGGGPDEETADKDGFVVLDCEEDEEDLVFEKQRTGSSATRATVNLADL 57
SD+EGG E +D DG + E + E+ + S+A R T+ L D+
Sbjct 858 GSDSEGGCDSEGGSDSDGGDFESVTPEHESRILEENVSSSTAERHTLILEDV 909
> At1g79970_1
Length=263
Score = 30.4 bits (67), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 26/99 (26%), Positives = 44/99 (44%), Gaps = 6/99 (6%)
Query 13 GPDEETADKDGFVVLDCEEDE---EDLVFEKQRTGSSATRATVNLADLVMEQLKKQTENK 69
GP+ +T G +LDC+ + ED+ ++ S +T N ++ E K++EN
Sbjct 163 GPNPKTTYIFGDCILDCDPKDLGKEDIEIHEEGDDSFSTEKPQNHREVSAE---KESENA 219
Query 70 DKSQEERVESSLSPKVVEVYTAMAPFLARYRSGKMPKAF 108
+ E L P + + +A L YRSG + F
Sbjct 220 GAEESCYEEEDLFPMAMPLNPNLAESLENYRSGVVAAMF 258
Lambda K H
0.319 0.132 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8734742214
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40