bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1085_orf1
Length=134
Score E
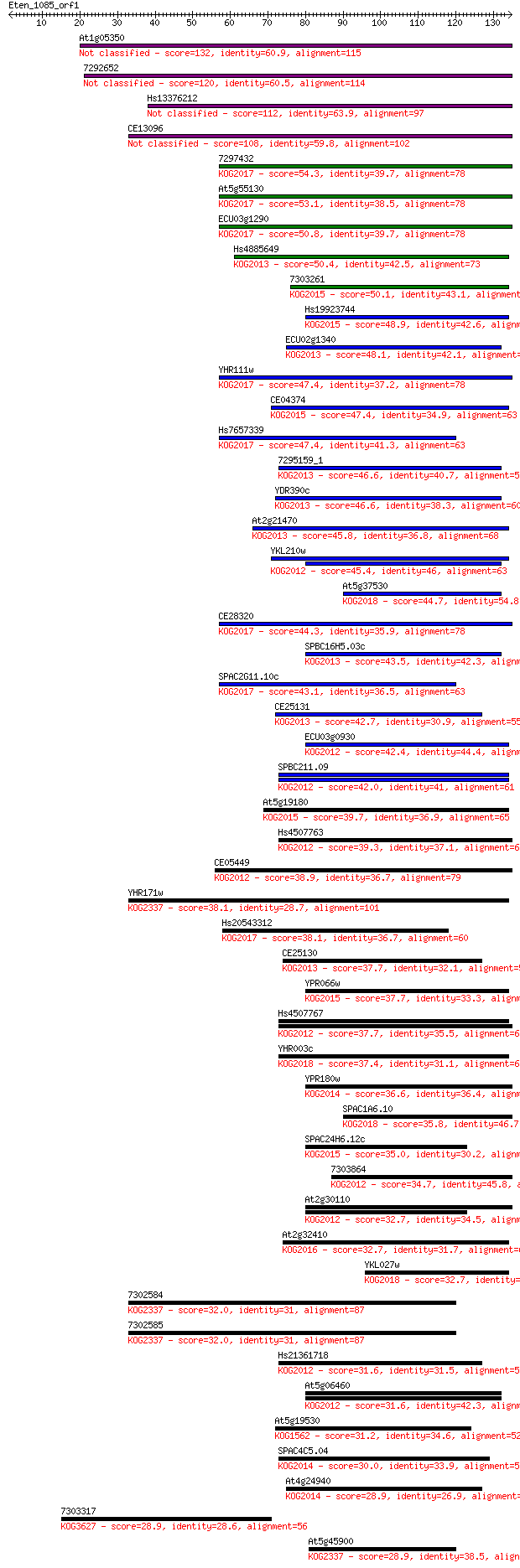
Sequences producing significant alignments: (Bits) Value
At1g05350 132 1e-31
7292652 120 7e-28
Hs13376212 112 2e-25
CE13096 108 3e-24
7297432 54.3 6e-08
At5g55130 53.1 2e-07
ECU03g1290 50.8 6e-07
Hs4885649 50.4 8e-07
7303261 50.1 1e-06
Hs19923744 48.9 3e-06
ECU02g1340 48.1 4e-06
YHR111w 47.4 8e-06
CE04374 47.4 8e-06
Hs7657339 47.4 8e-06
7295159_1 46.6 1e-05
YDR390c 46.6 1e-05
At2g21470 45.8 2e-05
YKL210w 45.4 3e-05
At5g37530 44.7 5e-05
CE28320 44.3 6e-05
SPBC16H5.03c 43.5 1e-04
SPAC2G11.10c 43.1 2e-04
CE25131 42.7 2e-04
ECU03g0930 42.4 3e-04
SPBC211.09 42.0 3e-04
At5g19180 39.7 0.002
Hs4507763 39.3 0.002
CE05449 38.9 0.003
YHR171w 38.1 0.004
Hs20543312 38.1 0.005
CE25130 37.7 0.006
YPR066w 37.7 0.007
Hs4507767 37.7 0.007
YHR003c 37.4 0.008
YPR180w 36.6 0.013
SPAC1A6.10 35.8 0.022
SPAC24H6.12c 35.0 0.040
7303864 34.7 0.049
At2g30110 32.7 0.18
At2g32410 32.7 0.20
YKL027w 32.7 0.21
7302584 32.0 0.33
7302585 32.0 0.37
Hs21361718 31.6 0.39
At5g06460 31.6 0.47
At5g19530 31.2 0.63
SPAC4C5.04 30.0 1.3
At4g24940 28.9 2.9
7303317 28.9 3.1
At5g45900 28.9 3.1
> At1g05350
Length=445
Score = 132 bits (333), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 70/120 (58%), Positives = 88/120 (73%), Gaps = 7/120 (5%)
Query 20 PAVLINLKMRKDAE-----SQTEGGLRPKVLSMSAEVRADNPYSRLMALQRMGVVRDYQA 74
PA LIN K+R E S+ R KV +S+EV NPYSRLMALQRMG+V +Y+
Sbjct 22 PA-LIN-KLRSHVENLATLSKCNPHRRSKVKELSSEVVDSNPYSRLMALQRMGIVDNYER 79
Query 75 ITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLFFTPKDVGMTKVEA 134
I + +V +VG+GGVGSVAAEML RCGIG+++L D+D+VEL+NMNRLFF P VGMTK +A
Sbjct 80 IREFSVAIVGIGGVGSVAAEMLTRCGIGRLLLYDYDTVELANMNRLFFRPDQVGMTKTDA 139
> 7292652
Length=404
Score = 120 bits (301), Expect = 7e-28, Method: Compositional matrix adjust.
Identities = 69/121 (57%), Positives = 88/121 (72%), Gaps = 7/121 (5%)
Query 21 AVLINLKMRKDAESQTEGGL-------RPKVLSMSAEVRADNPYSRLMALQRMGVVRDYQ 73
A++ +LK + E ++ GG+ R ++ MSAEV NPYSRLMALQRM +V+DY+
Sbjct 10 AIIADLKTELETEPKSSGGVASNSRLARDRIDRMSAEVVDSNPYSRLMALQRMNIVKDYE 69
Query 74 AITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLFFTPKDVGMTKVE 133
I KAV +VGVGGVGSV A+ML RCGIGK+IL D+D VEL+NMNRLFFTP G++KV
Sbjct 70 RIRYKAVAIVGVGGVGSVTADMLTRCGIGKLILFDYDKVELANMNRLFFTPDQAGLSKVA 129
Query 134 A 134
A
Sbjct 130 A 130
> Hs13376212
Length=404
Score = 112 bits (279), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 62/97 (63%), Positives = 76/97 (78%), Gaps = 0/97 (0%)
Query 38 GGLRPKVLSMSAEVRADNPYSRLMALQRMGVVRDYQAITKKAVLLVGVGGVGSVAAEMLV 97
GG R ++ MS+EV NPYSRLMAL+RMG+V DY+ I AV +VGVGGVGSV AEML
Sbjct 34 GGGRVRIEKMSSEVVDSNPYSRLMALKRMGIVSDYEKIRTFAVAIVGVGGVGSVTAEMLT 93
Query 98 RCGIGKIILLDFDSVELSNMNRLFFTPKDVGMTKVEA 134
RCGIGK++L D+D VEL+NMNRLFF P G++KV+A
Sbjct 94 RCGIGKLLLFDYDKVELANMNRLFFQPHQAGLSKVQA 130
> CE13096
Length=419
Score = 108 bits (270), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 61/102 (59%), Positives = 78/102 (76%), Gaps = 0/102 (0%)
Query 33 ESQTEGGLRPKVLSMSAEVRADNPYSRLMALQRMGVVRDYQAITKKAVLLVGVGGVGSVA 92
+ ++ R K+ +SAEV NPYSRLMALQRMG+V +Y+ I +K V +VGVGGVGSV
Sbjct 40 QPKSPAPYRQKIEKLSAEVVDSNPYSRLMALQRMGIVNEYERIREKTVAVVGVGGVGSVV 99
Query 93 AEMLVRCGIGKIILLDFDSVELSNMNRLFFTPKDVGMTKVEA 134
AEML RCGIGK+IL D+D VE++NMNRLF+ P G++KVEA
Sbjct 100 AEMLTRCGIGKLILFDYDKVEIANMNRLFYQPNQAGLSKVEA 141
> 7297432
Length=453
Score = 54.3 bits (129), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 31/79 (39%), Positives = 46/79 (58%), Gaps = 2/79 (2%)
Query 57 YSRLMALQRMGVVRDYQAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSN 116
YSR + L GV + + +VL+VG+GG+G AA+ L G G + L+D+D VE SN
Sbjct 72 YSRQLILPDFGVQGQLK-LKNSSVLIVGLGGLGCPAAQYLAAAGCGHLGLVDYDEVERSN 130
Query 117 MNRLFFTPKD-VGMTKVEA 134
+R +D GM+K E+
Sbjct 131 FHRQILHSEDRCGMSKAES 149
> At5g55130
Length=464
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 30/79 (37%), Positives = 47/79 (59%), Gaps = 2/79 (2%)
Query 57 YSRLMALQRMGVVRDYQAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSN 116
YSR + L V + K +VL++G GG+GS A L CG+G++ ++D D VEL+N
Sbjct 72 YSRQLLLPSFAV-EGQSNLLKSSVLVIGAGGLGSPALLYLAACGVGQLGIIDHDVVELNN 130
Query 117 MNR-LFFTPKDVGMTKVEA 134
M+R + T +G KV++
Sbjct 131 MHRQIIHTEAFIGHPKVKS 149
> ECU03g1290
Length=378
Score = 50.8 bits (120), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 31/79 (39%), Positives = 46/79 (58%), Gaps = 2/79 (2%)
Query 57 YSRLMALQRMGVVRDYQAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSN 116
YSR + + + V R +++ +L+VG GG+GS A L CG +I L+DFD VE+ N
Sbjct 15 YSRQIIVPGIHV-RGQKSLGDSGILVVGCGGLGSPAIMYLSSCGARRIGLVDFDKVEIHN 73
Query 117 MNR-LFFTPKDVGMTKVEA 134
+ R + +T DV KV A
Sbjct 74 LQRQVIYTEADVSQHKVVA 92
> Hs4885649
Length=640
Score = 50.4 bits (119), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 31/75 (41%), Positives = 46/75 (61%), Gaps = 3/75 (4%)
Query 61 MALQRMGVVRDY-QAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNR 119
MAL R G+ R+ +A+ VL+VG GG+G + LV G I L+D D++++SN+NR
Sbjct 1 MALSR-GLPRELAEAVAGGRVLVVGAGGIGCELLKNLVLTGFSHIDLIDLDTIDVSNLNR 59
Query 120 LF-FTPKDVGMTKVE 133
F F K VG +K +
Sbjct 60 QFLFQKKHVGRSKAQ 74
> 7303261
Length=450
Score = 50.1 bits (118), Expect = 1e-06, Method: Composition-based stats.
Identities = 25/59 (42%), Positives = 37/59 (62%), Gaps = 1/59 (1%)
Query 76 TKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLF-FTPKDVGMTKVE 133
TK VL++G GG+G + L G G + ++D D++ELSN+NR F F D+G +K E
Sbjct 47 TKCQVLIIGAGGLGCELLKDLALMGFGNLHVIDMDTIELSNLNRQFLFRRTDIGASKAE 105
> Hs19923744
Length=463
Score = 48.9 bits (115), Expect = 3e-06, Method: Composition-based stats.
Identities = 23/55 (41%), Positives = 36/55 (65%), Gaps = 1/55 (1%)
Query 80 VLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLF-FTPKDVGMTKVE 133
VL++G GG+G + L G +I ++D D++++SN+NR F F PKD+G K E
Sbjct 72 VLVIGAGGLGCELLKNLALSGFRQIHVIDMDTIDVSNLNRQFLFRPKDIGRPKAE 126
> ECU02g1340
Length=429
Score = 48.1 bits (113), Expect = 4e-06, Method: Composition-based stats.
Identities = 24/58 (41%), Positives = 37/58 (63%), Gaps = 1/58 (1%)
Query 75 ITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNR-LFFTPKDVGMTK 131
+T +L+VG GG+G ++L R + I L+D D+V+LSN+NR FF D+G +K
Sbjct 1 MTDGRILVVGCGGIGCELLKLLAREKLESITLVDSDTVDLSNLNRQFFFNRDDIGKSK 58
> YHR111w
Length=440
Score = 47.4 bits (111), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 29/79 (36%), Positives = 42/79 (53%), Gaps = 1/79 (1%)
Query 57 YSRLMALQRMGVVRDYQAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSN 116
Y R M ++ G V + VL+VG GG+G A L G+G+I ++D D VE SN
Sbjct 47 YGRQMIVEETGGVAGQVKLKNTKVLVVGAGGLGCPALPYLAGAGVGQIGIVDNDVVETSN 106
Query 117 MNR-LFFTPKDVGMTKVEA 134
++R + VGM K E+
Sbjct 107 LHRQVLHDSSRVGMLKCES 125
> CE04374
Length=437
Score = 47.4 bits (111), Expect = 8e-06, Method: Composition-based stats.
Identities = 22/64 (34%), Positives = 39/64 (60%), Gaps = 1/64 (1%)
Query 71 DYQAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLF-FTPKDVGM 129
+++A+ +L++G GG+G + L G I ++D D++++SN+NR F F DVG
Sbjct 36 NFEALQNTKILVIGAGGLGCELLKNLALSGFRTIEVIDMDTIDVSNLNRQFLFRESDVGK 95
Query 130 TKVE 133
+K E
Sbjct 96 SKAE 99
> Hs7657339
Length=460
Score = 47.4 bits (111), Expect = 8e-06, Method: Composition-based stats.
Identities = 26/63 (41%), Positives = 39/63 (61%), Gaps = 1/63 (1%)
Query 57 YSRLMALQRMGVVRDYQAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSN 116
YSR + L +GV + T VL+VG GG+G A+ L G+G++ L+D+D VE+SN
Sbjct 63 YSRQLVLPELGVHGQLRLGTA-CVLIVGCGGLGCPLAQYLAAAGVGRLGLVDYDVVEMSN 121
Query 117 MNR 119
+ R
Sbjct 122 LAR 124
> 7295159_1
Length=731
Score = 46.6 bits (109), Expect = 1e-05, Method: Composition-based stats.
Identities = 24/60 (40%), Positives = 37/60 (61%), Gaps = 1/60 (1%)
Query 73 QAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLF-FTPKDVGMTK 131
+ + K VL+VG GG+G + LV G I ++D D+++LSN+NR F F + VG +K
Sbjct 15 ELVKKSKVLVVGAGGIGCEVLKNLVLSGFTDIEIIDLDTIDLSNLNRQFLFHREHVGKSK 74
> YDR390c
Length=636
Score = 46.6 bits (109), Expect = 1e-05, Method: Composition-based stats.
Identities = 23/61 (37%), Positives = 37/61 (60%), Gaps = 1/61 (1%)
Query 72 YQAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLF-FTPKDVGMT 130
Y+ + LLVG GG+GS + ++ G+I ++D D+++LSN+NR F F KD+
Sbjct 16 YKKLRSSRCLLVGAGGIGSELLKDIILMEFGEIHIVDLDTIDLSNLNRQFLFRQKDIKQP 75
Query 131 K 131
K
Sbjct 76 K 76
> At2g21470
Length=700
Score = 45.8 bits (107), Expect = 2e-05, Method: Composition-based stats.
Identities = 25/69 (36%), Positives = 37/69 (53%), Gaps = 1/69 (1%)
Query 66 MGVVRDYQAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLF-FTP 124
M + AI VL+VG GG+G + L G I ++D D++E+SN+NR F F
Sbjct 1 MATQQQQSAIKGAKVLMVGAGGIGCELLKTLALSGFEDIHIIDMDTIEVSNLNRQFLFRR 60
Query 125 KDVGMTKVE 133
VG +K +
Sbjct 61 SHVGQSKAK 69
> YKL210w
Length=1024
Score = 45.4 bits (106), Expect = 3e-05, Method: Composition-based stats.
Identities = 29/70 (41%), Positives = 37/70 (52%), Gaps = 7/70 (10%)
Query 71 DYQA-ITKKAVLLVGVGGVGSVAAEMLVRCGIGK-----IILLDFDSVELSNMNRLF-FT 123
D+Q I V LVG G +G + G+G I++ D DS+E SN+NR F F
Sbjct 427 DFQKKIANSKVFLVGSGAIGCEMLKNWALLGLGSGSDGYIVVTDNDSIEKSNLNRQFLFR 486
Query 124 PKDVGMTKVE 133
PKDVG K E
Sbjct 487 PKDVGKNKSE 496
Score = 34.7 bits (78), Expect = 0.050, Method: Composition-based stats.
Identities = 17/53 (32%), Positives = 34/53 (64%), Gaps = 1/53 (1%)
Query 80 VLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLFF-TPKDVGMTK 131
VL++G+ G+G A+ +V G+ + + D + V+L++++ FF T KD+G +
Sbjct 39 VLILGLKGLGVEIAKNVVLAGVKSMTVFDPEPVQLADLSTQFFLTEKDIGQKR 91
> At5g37530
Length=414
Score = 44.7 bits (104), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 23/43 (53%), Positives = 31/43 (72%), Gaps = 1/43 (2%)
Query 90 SVAAEMLVRCGIGKIILLDFDSVELSNMNR-LFFTPKDVGMTK 131
S AA ML+R G+GK++L+DFD V LS++NR T DVG+ K
Sbjct 63 SHAASMLLRSGVGKLLLVDFDQVSLSSLNRHAVATRADVGIPK 105
> CE28320
Length=402
Score = 44.3 bits (103), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 28/79 (35%), Positives = 43/79 (54%), Gaps = 2/79 (2%)
Query 57 YSRLMALQRMGVVRDYQAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSN 116
YSR + + GV + + VL+VG GG+G A L GIG I ++D+D + L N
Sbjct 18 YSRQLLVDDFGV-SGQKNLKNLNVLIVGAGGLGCPVATYLGAAGIGTIGIVDYDHISLDN 76
Query 117 MNRLFFTPKD-VGMTKVEA 134
++R +D VG +K +A
Sbjct 77 LHRQVAYKEDQVGKSKAQA 95
> SPBC16H5.03c
Length=628
Score = 43.5 bits (101), Expect = 1e-04, Method: Composition-based stats.
Identities = 22/53 (41%), Positives = 34/53 (64%), Gaps = 1/53 (1%)
Query 80 VLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLF-FTPKDVGMTK 131
VLLVG GG+G + L+ G+ ++ ++D D+++LSN+NR F F K V K
Sbjct 28 VLLVGAGGIGCELLKNLLMSGVKEVHIIDLDTIDLSNLNRQFLFRKKHVKQPK 80
> SPAC2G11.10c
Length=401
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 23/63 (36%), Positives = 39/63 (61%), Gaps = 1/63 (1%)
Query 57 YSRLMALQRMGVVRDYQAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSN 116
Y R M L +G+ ++ + +VL++G GG+G A + LV GIG + ++D D V+ SN
Sbjct 24 YGRQMLLSEIGLPGQL-SLKRSSVLVIGAGGLGCPAMQYLVAAGIGTLGIMDGDVVDKSN 82
Query 117 MNR 119
++R
Sbjct 83 LHR 85
> CE25131
Length=438
Score = 42.7 bits (99), Expect = 2e-04, Method: Composition-based stats.
Identities = 17/55 (30%), Positives = 34/55 (61%), Gaps = 0/55 (0%)
Query 72 YQAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLFFTPKD 126
++ I + +L++G GG+G + L G K+ ++D D++++SN+NR F K+
Sbjct 8 HEKIVQSKILVIGAGGIGCELLKNLAVTGFRKVHVIDLDTIDISNLNRQFLFRKE 62
> ECU03g0930
Length=991
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 24/58 (41%), Positives = 33/58 (56%), Gaps = 4/58 (6%)
Query 80 VLLVGVGGVGSVAAEMLVRCGIG---KIILLDFDSVELSNMNRLF-FTPKDVGMTKVE 133
V +VG G +G + +V CGIG +I + D D++E SN+NR F F DV K E
Sbjct 416 VFMVGAGAIGCEHLKNMVMCGIGSNGRISVTDMDAIEQSNLNRQFLFRSGDVSSMKAE 473
> SPBC211.09
Length=644
Score = 42.0 bits (97), Expect = 3e-04, Method: Composition-based stats.
Identities = 25/67 (37%), Positives = 34/67 (50%), Gaps = 6/67 (8%)
Query 73 QAITKKAVLLVGVGGVGSVAAEMLVRCGI-----GKIILLDFDSVELSNMNRLF-FTPKD 126
+ I + LVG G +G + G+ G I + D DS+E SN+NR F F P+D
Sbjct 423 EKIASLSTFLVGAGAIGCEMLKNWAMMGVATGESGHISVTDMDSIEKSNLNRQFLFRPRD 482
Query 127 VGMTKVE 133
VG K E
Sbjct 483 VGKLKSE 489
Score = 27.7 bits (60), Expect = 5.9, Method: Composition-based stats.
Identities = 13/62 (20%), Positives = 33/62 (53%), Gaps = 1/62 (1%)
Query 73 QAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNM-NRLFFTPKDVGMTK 131
+ +++ VL++G G+G A+ + G+ + L D + ++ ++ F T D+G+ +
Sbjct 33 KQMSQSNVLIIGCKGLGVEIAKNVCLAGVKSVTLYDPQPTRIEDLSSQYFLTEDDIGVPR 92
Query 132 VE 133
+
Sbjct 93 AK 94
> At5g19180
Length=477
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 24/66 (36%), Positives = 37/66 (56%), Gaps = 6/66 (9%)
Query 69 VRDYQAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLF-FTPKDV 127
+RDY I L++G GG+G + L G + ++D D +E++N+NR F F +DV
Sbjct 43 IRDYVRI-----LVIGAGGLGCELLKDLALSGFRNLEVIDMDRIEVTNLNRQFLFRIEDV 97
Query 128 GMTKVE 133
G K E
Sbjct 98 GKPKAE 103
> Hs4507763
Length=1058
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 35/68 (51%), Gaps = 6/68 (8%)
Query 73 QAITKKAVLLVGVGGVGSVAAEMLVRCGIG-----KIILLDFDSVELSNMNRLF-FTPKD 126
+ + K+ LVG G +G + G+G +II+ D D++E SN+NR F F P D
Sbjct 464 EKLGKQKYFLVGAGAIGCELLKNFAMIGLGCGEGGEIIVTDMDTIEKSNLNRQFLFRPWD 523
Query 127 VGMTKVEA 134
V K +
Sbjct 524 VTKLKSDT 531
> CE05449
Length=1113
Score = 38.9 bits (89), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 29/88 (32%), Positives = 39/88 (44%), Gaps = 26/88 (29%)
Query 56 PYSRLMALQRMGVVRDYQAITKKAVLLVGVGGVGSVAAEML-------VRCGIGKII-LL 107
PY + QR VV G G++ E+L V CG G +I +
Sbjct 510 PYQECLFRQRWFVV-----------------GAGAIGCELLKNLSMMGVACGEGGLIKIT 552
Query 108 DFDSVELSNMNRLF-FTPKDVGMTKVEA 134
D D +E+SN+NR F F +DVG K E
Sbjct 553 DMDQIEISNLNRQFLFRRRDVGGKKSEC 580
> YHR171w
Length=630
Score = 38.1 bits (87), Expect = 0.004, Method: Composition-based stats.
Identities = 29/108 (26%), Positives = 50/108 (46%), Gaps = 7/108 (6%)
Query 33 ESQTEGGLRPKVLSMSAEVR----ADNPYSRLMALQRMGVVRDYQ--AITKKAVLLVGVG 86
E +G L P+V+ +S+ + AD + L + ++ D I VLL+G G
Sbjct 274 ERNVQGKLAPRVVDLSSLLDPLKIADQSVDLNLKLMKWRILPDLNLDIIKNTKVLLLGAG 333
Query 87 GVGSVAAEMLVRCGIGKIILLDFDSVELSN-MNRLFFTPKDVGMTKVE 133
+G + L+ G+ KI +D +V SN + + + +D G K E
Sbjct 334 TLGCYVSRALIAWGVRKITFVDNGTVSYSNPVRQALYNFEDCGKPKAE 381
> Hs20543312
Length=306
Score = 38.1 bits (87), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 22/60 (36%), Positives = 34/60 (56%), Gaps = 8/60 (13%)
Query 58 SRLMALQRMGVVRDYQAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNM 117
SR + L +GV R VL+VG GG+ A+ L +G++ L+D+D VE+SN+
Sbjct 73 SRQLVLPELGVHRH--------VLVVGCGGLRCPLAQYLAAASVGRLGLVDYDVVEMSNL 124
> CE25130
Length=456
Score = 37.7 bits (86), Expect = 0.006, Method: Composition-based stats.
Identities = 17/54 (31%), Positives = 34/54 (62%), Gaps = 1/54 (1%)
Query 74 AITKKAVL-LVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLFFTPKD 126
+I+K ++ ++G GG+G + L G K+ ++D D++++SN+NR F K+
Sbjct 27 SISKISIFQVIGAGGIGCELLKNLAVTGFRKVHVIDLDTIDISNLNRQFLFRKE 80
> YPR066w
Length=299
Score = 37.7 bits (86), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 18/56 (32%), Positives = 36/56 (64%), Gaps = 2/56 (3%)
Query 80 VLLVGVGGVGS-VAAEMLVRCGIGKIILLDFDSVELSNMNRLF-FTPKDVGMTKVE 133
+L++G GG+G + + + + ++ ++D D++EL+N+NR F F KD+G K +
Sbjct 5 ILVLGAGGLGCEILKNLTMLSFVKQVHIVDIDTIELTNLNRQFLFCDKDIGKPKAQ 60
> Hs4507767
Length=1011
Score = 37.7 bits (86), Expect = 0.007, Method: Composition-based stats.
Identities = 21/67 (31%), Positives = 36/67 (53%), Gaps = 6/67 (8%)
Query 73 QAITKKAVLLVGVGGVGSVAAEMLVRCGIGK-----IILLDFDSVELSNMNRLF-FTPKD 126
+ + ++ LLVG G +G ++ G+G + ++D D +E SN++R F F +D
Sbjct 427 EKLRRQHYLLVGAGAIGCELLKVFALVGLGAGNSGGLTVVDMDHIERSNLSRQFLFRSQD 486
Query 127 VGMTKVE 133
VG K E
Sbjct 487 VGRPKAE 493
Score = 28.5 bits (62), Expect = 4.1, Method: Composition-based stats.
Identities = 19/63 (30%), Positives = 33/63 (52%), Gaps = 1/63 (1%)
Query 73 QAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLF-FTPKDVGMTK 131
Q I VL+ G+ G+G+ A+ LV G+G + L D S++ F + +D+ ++
Sbjct 28 QRIQGARVLVSGLQGLGAEVAKNLVLMGVGSLTLHDPHPTCWSDLAAQFLLSEQDLERSR 87
Query 132 VEA 134
EA
Sbjct 88 AEA 90
> YHR003c
Length=429
Score = 37.4 bits (85), Expect = 0.008, Method: Composition-based stats.
Identities = 19/62 (30%), Positives = 37/62 (59%), Gaps = 1/62 (1%)
Query 73 QAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMN-RLFFTPKDVGMTK 131
+ I ++ +++VG G VGS ML+R G KI+++D +++ + ++N D+G K
Sbjct 70 RKIKEQYIVIVGAGEVGSWVCTMLIRSGCQKIMIIDPENISIDSLNTHCCAVLSDIGKPK 129
Query 132 VE 133
V+
Sbjct 130 VQ 131
> YPR180w
Length=347
Score = 36.6 bits (83), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 20/56 (35%), Positives = 32/56 (57%), Gaps = 1/56 (1%)
Query 80 VLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLFFT-PKDVGMTKVEA 134
VLL+ +G +GS + +V GIG + +LD V ++ FF +DVG K++A
Sbjct 36 VLLINLGAIGSEITKSIVLSGIGHLTILDGHMVTEEDLGSQFFIGSEDVGQWKIDA 91
> SPAC1A6.10
Length=485
Score = 35.8 bits (81), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 21/46 (45%), Positives = 27/46 (58%), Gaps = 1/46 (2%)
Query 90 SVAAEMLVRCGIGKIILLDFDSVELSNMNRLFF-TPKDVGMTKVEA 134
S ML R G+ KI ++DFD V LS++NR T +DVG K A
Sbjct 139 SWVINMLARSGVQKIRIVDFDQVSLSSLNRHSIATLQDVGTPKTLA 184
> SPAC24H6.12c
Length=444
Score = 35.0 bits (79), Expect = 0.040, Method: Composition-based stats.
Identities = 13/43 (30%), Positives = 27/43 (62%), Gaps = 0/43 (0%)
Query 80 VLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLFF 122
+L++G GG+G + L G + ++D D+++++N+NR F
Sbjct 47 ILIIGAGGLGCEILKDLALSGFRDLSVIDMDTIDITNLNRQFL 89
> 7303864
Length=1008
Score = 34.7 bits (78), Expect = 0.049, Method: Composition-based stats.
Identities = 22/60 (36%), Positives = 31/60 (51%), Gaps = 12/60 (20%)
Query 87 GVGSVAAEMLVRCGI-------GKIILLDFDSVELSNMNRLF-FTPKDV----GMTKVEA 134
G G++ E+L G+ G+I + D D +E SN+NR F F P DV MT +A
Sbjct 428 GAGAIGCELLKNFGMLGLGTGNGQIFVTDMDLIEKSNLNRQFLFRPHDVQKPKSMTAADA 487
> At2g30110
Length=1080
Score = 32.7 bits (73), Expect = 0.18, Method: Composition-based stats.
Identities = 19/56 (33%), Positives = 33/56 (58%), Gaps = 1/56 (1%)
Query 80 VLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLF-FTPKDVGMTKVEA 134
VL+ G+ G+G+ A+ L+ G+ + L D VEL +++ F F+ DVG + +A
Sbjct 98 VLISGMHGLGAEIAKNLILAGVKSVTLHDERVVELWDLSSNFVFSEDDVGKNRADA 153
Score = 27.3 bits (59), Expect = 7.8, Method: Composition-based stats.
Identities = 19/48 (39%), Positives = 25/48 (52%), Gaps = 5/48 (10%)
Query 80 VLLVGVGGVG----SVAAEMLVRCGI-GKIILLDFDSVELSNMNRLFF 122
V VG G +G A M V CG GK+ + D D +E SN++R F
Sbjct 495 VFTVGSGALGCEFLKNLALMGVSCGSQGKLTVTDDDIIEKSNLSRQFL 542
> At2g32410
Length=523
Score = 32.7 bits (73), Expect = 0.20, Method: Composition-based stats.
Identities = 19/61 (31%), Positives = 32/61 (52%), Gaps = 1/61 (1%)
Query 74 AITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNM-NRLFFTPKDVGMTKV 132
A+ ++ L+ G GS A + LV GIG I ++D VE+ ++ N K VG ++
Sbjct 22 ALETASICLLNCGPTGSEALKNLVIGGIGSITIVDGSKVEIGDLGNNFMVDAKSVGQSRA 81
Query 133 E 133
+
Sbjct 82 K 82
> YKL027w
Length=447
Score = 32.7 bits (73), Expect = 0.21, Method: Composition-based stats.
Identities = 20/39 (51%), Positives = 24/39 (61%), Gaps = 1/39 (2%)
Query 96 LVRCGIGKIILLDFDSVELSNMNRLFFTP-KDVGMTKVE 133
LVR G KI ++DFD V LS++NR DVG KVE
Sbjct 105 LVRSGCRKIRVVDFDQVSLSSLNRHSCAILNDVGTPKVE 143
> 7302584
Length=684
Score = 32.0 bits (71), Expect = 0.33, Method: Composition-based stats.
Identities = 27/93 (29%), Positives = 41/93 (44%), Gaps = 6/93 (6%)
Query 33 ESQTEGGLRPKVL----SMSAEVRADNPYSRLMALQRMGVVRDY--QAITKKAVLLVGVG 86
E G + P+++ SM A+N + + L + +V D + I++ LL G G
Sbjct 289 ELNKNGKMGPRMVCMRDSMDPAKLAENSVNLNLKLMKWRLVPDLNLEIISQTKCLLFGAG 348
Query 87 GVGSVAAEMLVRCGIGKIILLDFDSVELSNMNR 119
+G A L+ G I LLD V SN R
Sbjct 349 TLGCAVARNLLSWGFKHITLLDSGKVGFSNPVR 381
> 7302585
Length=510
Score = 32.0 bits (71), Expect = 0.37, Method: Composition-based stats.
Identities = 27/93 (29%), Positives = 41/93 (44%), Gaps = 6/93 (6%)
Query 33 ESQTEGGLRPKVL----SMSAEVRADNPYSRLMALQRMGVVRDY--QAITKKAVLLVGVG 86
E G + P+++ SM A+N + + L + +V D + I++ LL G G
Sbjct 115 ELNKNGKMGPRMVCMRDSMDPAKLAENSVNLNLKLMKWRLVPDLNLEIISQTKCLLFGAG 174
Query 87 GVGSVAAEMLVRCGIGKIILLDFDSVELSNMNR 119
+G A L+ G I LLD V SN R
Sbjct 175 TLGCAVARNLLSWGFKHITLLDSGKVGFSNPVR 207
> Hs21361718
Length=1052
Score = 31.6 bits (70), Expect = 0.39, Method: Composition-based stats.
Identities = 17/54 (31%), Positives = 28/54 (51%), Gaps = 0/54 (0%)
Query 73 QAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLFFTPKD 126
Q + K V L G+GG+G A+ LV GI + + D + + ++ FF +D
Sbjct 57 QKMAKSHVFLSGMGGLGLEIAKNLVLAGIKAVTIHDTEKCQAWDLGTNFFLSED 110
> At5g06460
Length=1077
Score = 31.6 bits (70), Expect = 0.47, Method: Compositional matrix adjust.
Identities = 22/58 (37%), Positives = 31/58 (53%), Gaps = 6/58 (10%)
Query 80 VLLVGVGGVGSVA----AEMLVRCGI-GKIILLDFDSVELSNMNRLF-FTPKDVGMTK 131
V +VG G +G A M V CG GK+ + D D +E SN++R F F ++G K
Sbjct 492 VFVVGAGALGCEFLKNLALMGVSCGTQGKLTVTDDDVIEKSNLSRQFLFRDWNIGQAK 549
Score = 31.2 bits (69), Expect = 0.54, Method: Compositional matrix adjust.
Identities = 17/53 (32%), Positives = 32/53 (60%), Gaps = 1/53 (1%)
Query 80 VLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLF-FTPKDVGMTK 131
VL+ G+ G+G A+ ++ G+ + L D + VEL +++ F FT +D+G +
Sbjct 95 VLISGMQGLGVEIAKNIILAGVKSVTLHDENVVELWDLSSNFVFTEEDIGKNR 147
> At5g19530
Length=340
Score = 31.2 bits (69), Expect = 0.63, Method: Compositional matrix adjust.
Identities = 18/52 (34%), Positives = 28/52 (53%), Gaps = 2/52 (3%)
Query 72 YQAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLFFT 123
+ + K V ++G GG GS A E+L I K+++ D D E+ + R F T
Sbjct 103 FHPNSPKTVFIMG-GGEGSAAREILKHTTIEKVVMCDIDQ-EVVDFCRRFLT 152
> SPAC4C5.04
Length=307
Score = 30.0 bits (66), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 31/57 (54%), Gaps = 1/57 (1%)
Query 73 QAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLFFT-PKDVG 128
QA+ + VLL+ + + A+ LV GIGK+ +LD +V ++ FF D+G
Sbjct 27 QALKQSRVLLITASPLANEIAKNLVLSGIGKLCVLDSMTVYEKDVEEQFFIEASDIG 83
> At4g24940
Length=319
Score = 28.9 bits (63), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 14/52 (26%), Positives = 26/52 (50%), Gaps = 0/52 (0%)
Query 75 ITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLFFTPKD 126
+TK +L+ G+ G + + +V G+G + L+D + +N F P D
Sbjct 29 LTKAHILVSGIKGTVAEFCKNIVLAGVGSVTLMDDRLANMEALNANFLIPPD 80
> 7303317
Length=260
Score = 28.9 bits (63), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 16/56 (28%), Positives = 28/56 (50%), Gaps = 4/56 (7%)
Query 15 HLQARPAVLINLKMRKDAESQTEGGLRPKVLSMSAEVRADNPYSRLMALQRMGVVR 70
H P + NL++R + +T GG+ +V A ++A Y+ + +GVVR
Sbjct 75 HCLDTPTTVSNLRIRAGSNKRTYGGVLVEV----AAIKAHEAYNSNSKINDIGVVR 126
> At5g45900
Length=678
Score = 28.9 bits (63), Expect = 3.1, Method: Composition-based stats.
Identities = 15/39 (38%), Positives = 21/39 (53%), Gaps = 0/39 (0%)
Query 81 LLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNR 119
LL+G G +G A L+ GI I +D+ V +SN R
Sbjct 342 LLLGAGTLGCQVARTLMGWGIRNITFVDYGKVAMSNPVR 380
Lambda K H
0.321 0.137 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1393086308
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40